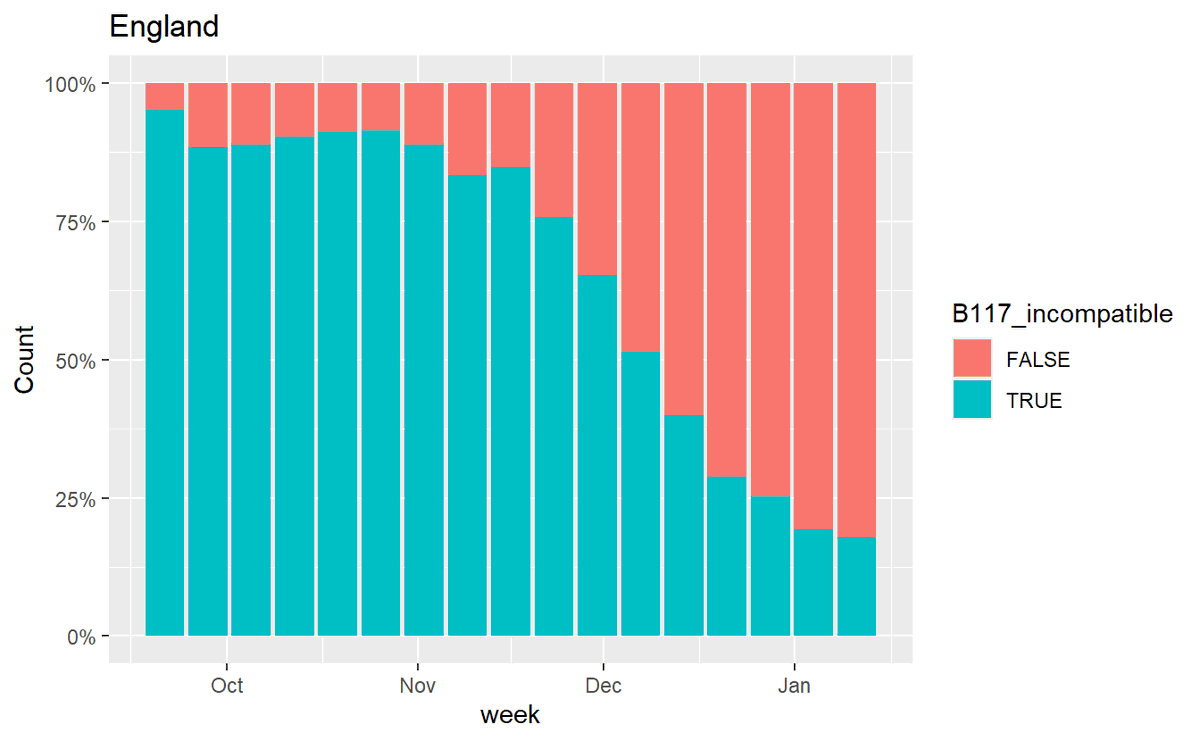
1/ The ONS infection survey has come out, and the graph below has led some people to speculate that B.1.1.7 isn't more transmissible any more.
Unfortunately I don't think we can conclude that. Here is why [1/n]
Unfortunately I don't think we can conclude that. Here is why [1/n]
2/ The apparent clincher here is that this would suggest that new-variant-compatible cases are falling *as a proportion of cases*. It was the increase in B.1.1.7 as a proportion of genomes in multiple areas that suggested its increased transmissibility.
3/ But what do the ONS mean by new variant compatible? Each TaqPath test amplifies 3 targets: S, N and OR. Each of these can give a positive or negative result. So any TaqPath test could give one of 7 results:
- OR+N+S
- OR+N
- OR+S
- N+S
- just N
- just S
- just OR
- OR+N+S
- OR+N
- OR+S
- N+S
- just N
- just S
- just OR
4/ The ONS definition of new-variant-compatible is when just the S gene drops out, and you get OR+N. But we know that amplicons can drop out randomly, esp. if you have few viruses in a sample. So the combinations technically compatible with B.1.1.7 are:
- OR+N
- just N
- just OR
- OR+N
- just N
- just OR
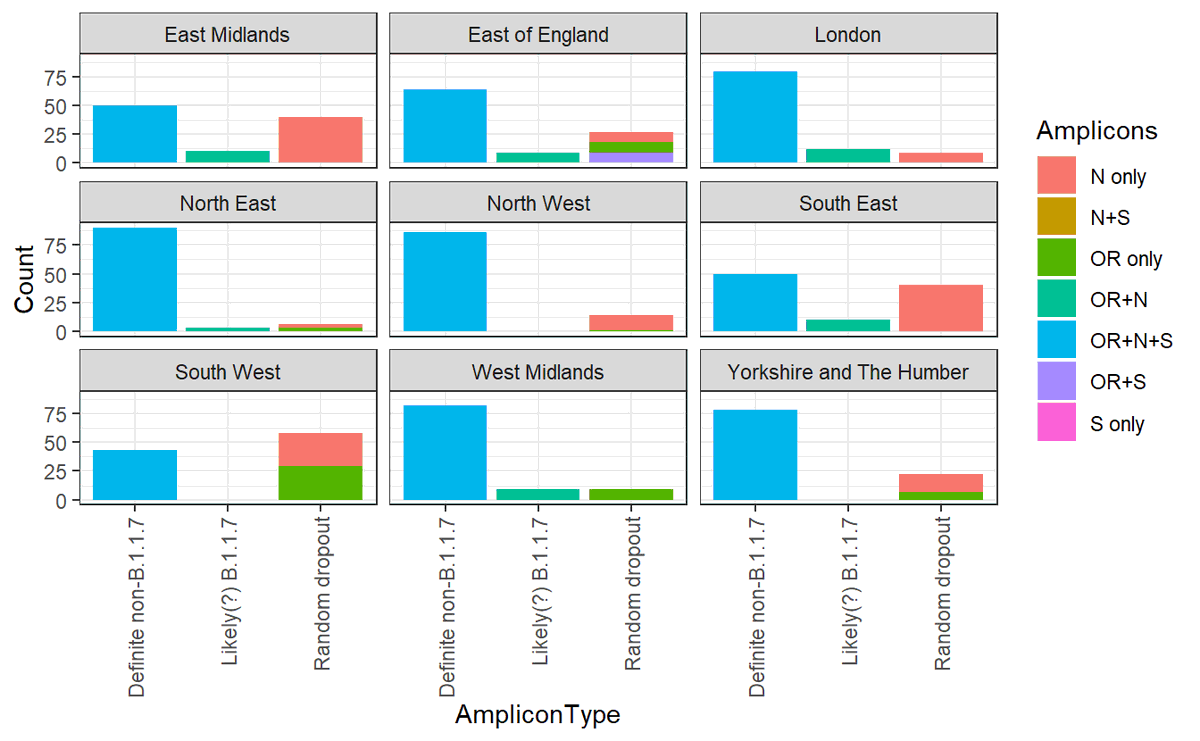
5/ I had a look at ONS data from September (before B.1.1.7 was widespread) to show that this random dropout occurs, and correlates with the general levels of viral load in a region
High Ct values mean low amounts of virus per sample, and lead to more of this random dropout
High Ct values mean low amounts of virus per sample, and lead to more of this random dropout
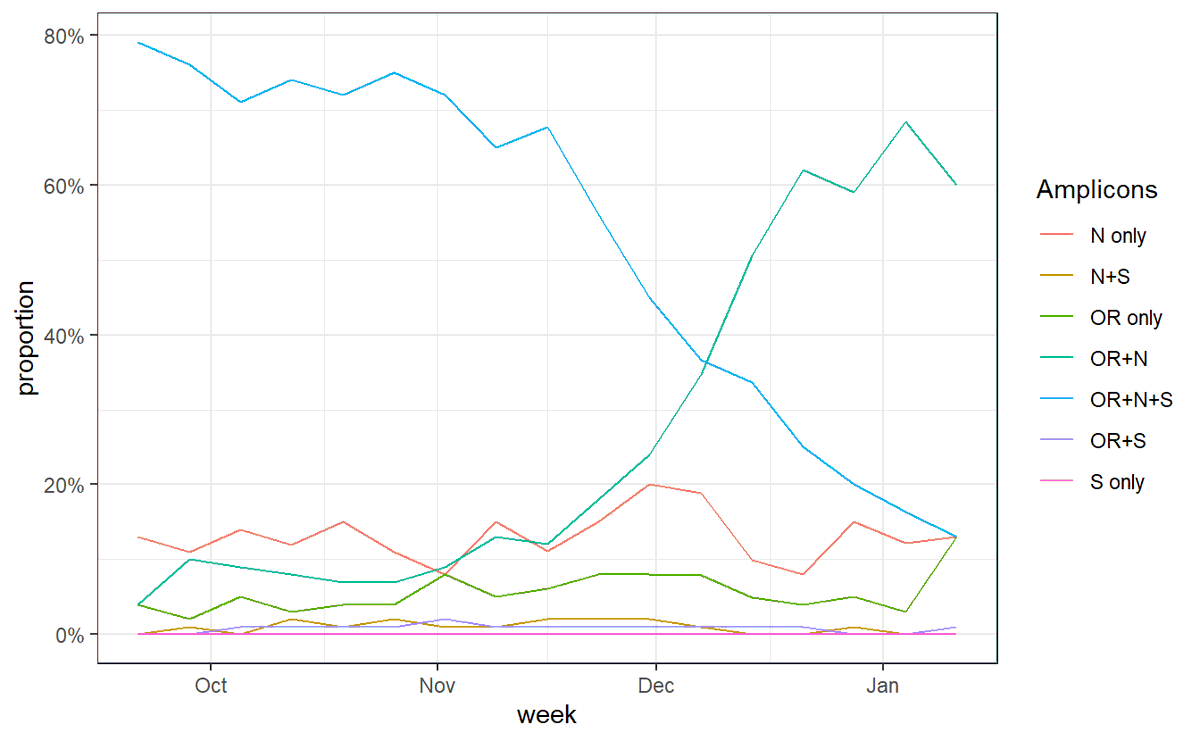
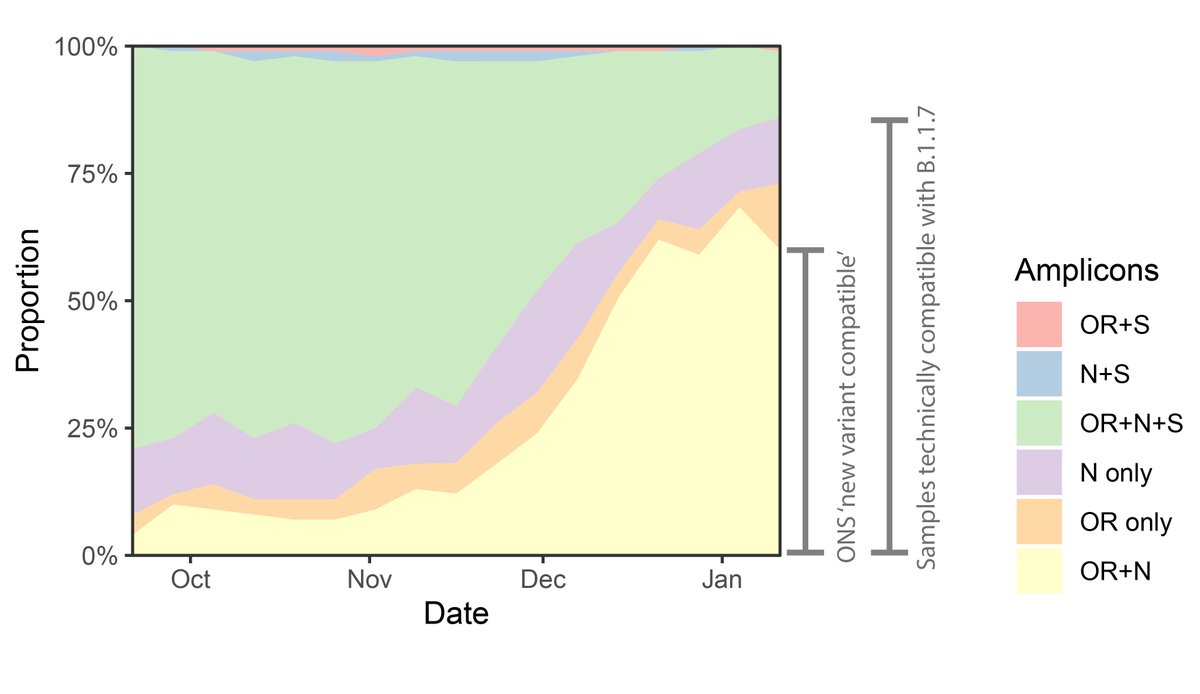
6/ So what's happened recently in England? If we look at all of these different combinations over time we can see that OR+N (which is how the ONS defines 'new-variant-compatible') has indeed flatlined or fallen lately.
7/ But it hasn't been replaced by OR+N+S (which is essentially incompatible with B.1.1.7), it's been replaced by N only and OR only, which are completely compatible with being either B.1.1.7.
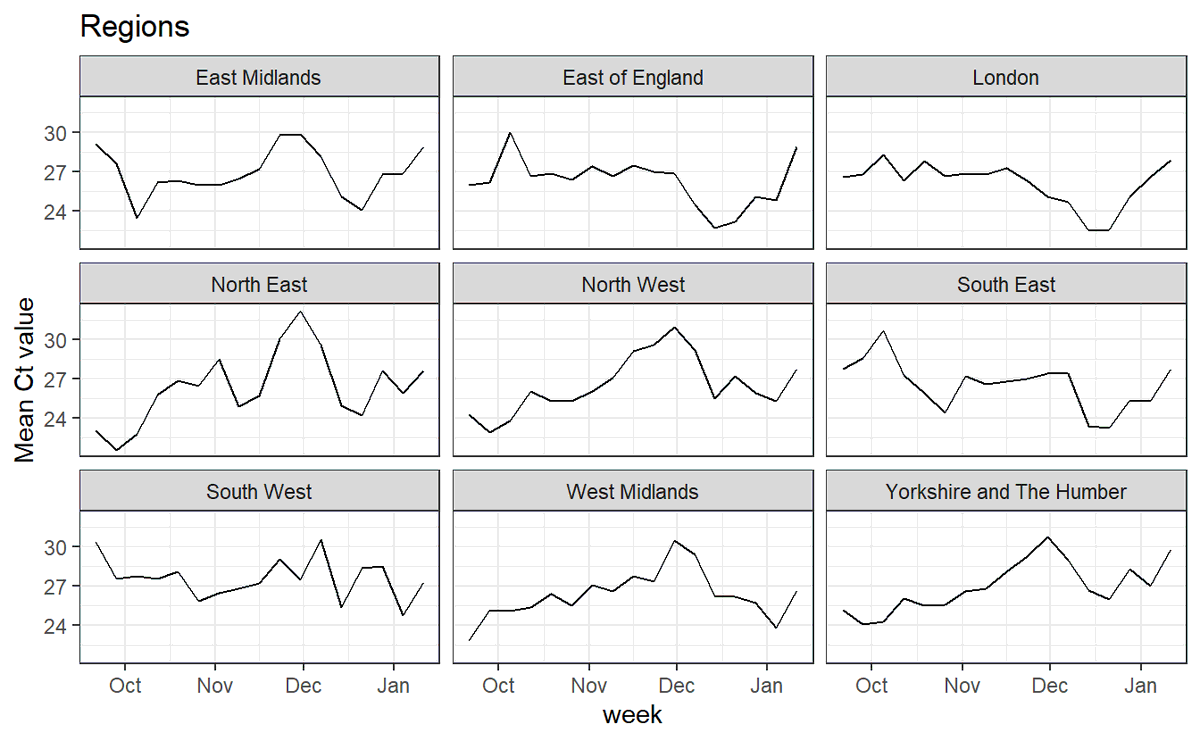
8/ And we can see that mean Ct has risen in this recent period, explaining the change from OR+N to OR only or N only (random dropout of one or other amplicon)
9/ Finally (for now), it's really complicated. Random dropout of S could lead WT viruses to present as "OR+N" (new-variant-compatible). SGTL could lead a proportion B.1.1.7 to present as "OR+N+S", creating an upper limit on apparent B.1.1.7 prevalence..
10/ ..Other variants might have come along that are genuinely as transmissible as B.1.1.7. Other variants might have appeared which always show N or OR dropout due to their genomes. No analysis method is going to be perfect with this type of data, it's lucky we have genomes too.
11/ You can check my workings in this work-in-progress blog post: https://theo.io/post/2021-01-22-ons-data/
12/ Something I should have mentioned in the thread is why Ct values are rising (number of viruses in samples falling). It's because we're controlling new infections better, so a higher proportion of the infections we identify are from people who were infected some time ago.
13/ The ONS data are available here, in sheets 6a and 6b. https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/coronaviruscovid19infectionsurveypilot/24december2020/relateddata It is to their very great credit that this level of detail is available.
(My own view is that line-level Ct data would be helpful here, and without demographic data wouldn't compromise privacy)
(My own view is that line-level Ct data would be helpful here, and without demographic data wouldn't compromise privacy)
14/ Just made this, which I think is an easier way to look at these different ways of considering B.1.1.7 compatibility.

 Read on Twitter
Read on Twitter![1/ The ONS infection survey has come out, and the graph below has led some people to speculate that B.1.1.7 isn't more transmissible any more. Unfortunately I don't think we can conclude that. Here is why [1/n] 1/ The ONS infection survey has come out, and the graph below has led some people to speculate that B.1.1.7 isn't more transmissible any more. Unfortunately I don't think we can conclude that. Here is why [1/n]](https://pbs.twimg.com/media/EsWfbM8W8AI9GAS.png)