Our analyses on the emergence and transmission of novel SARS-CoV-2 variants with deletions H69/V70 in Spike protein.
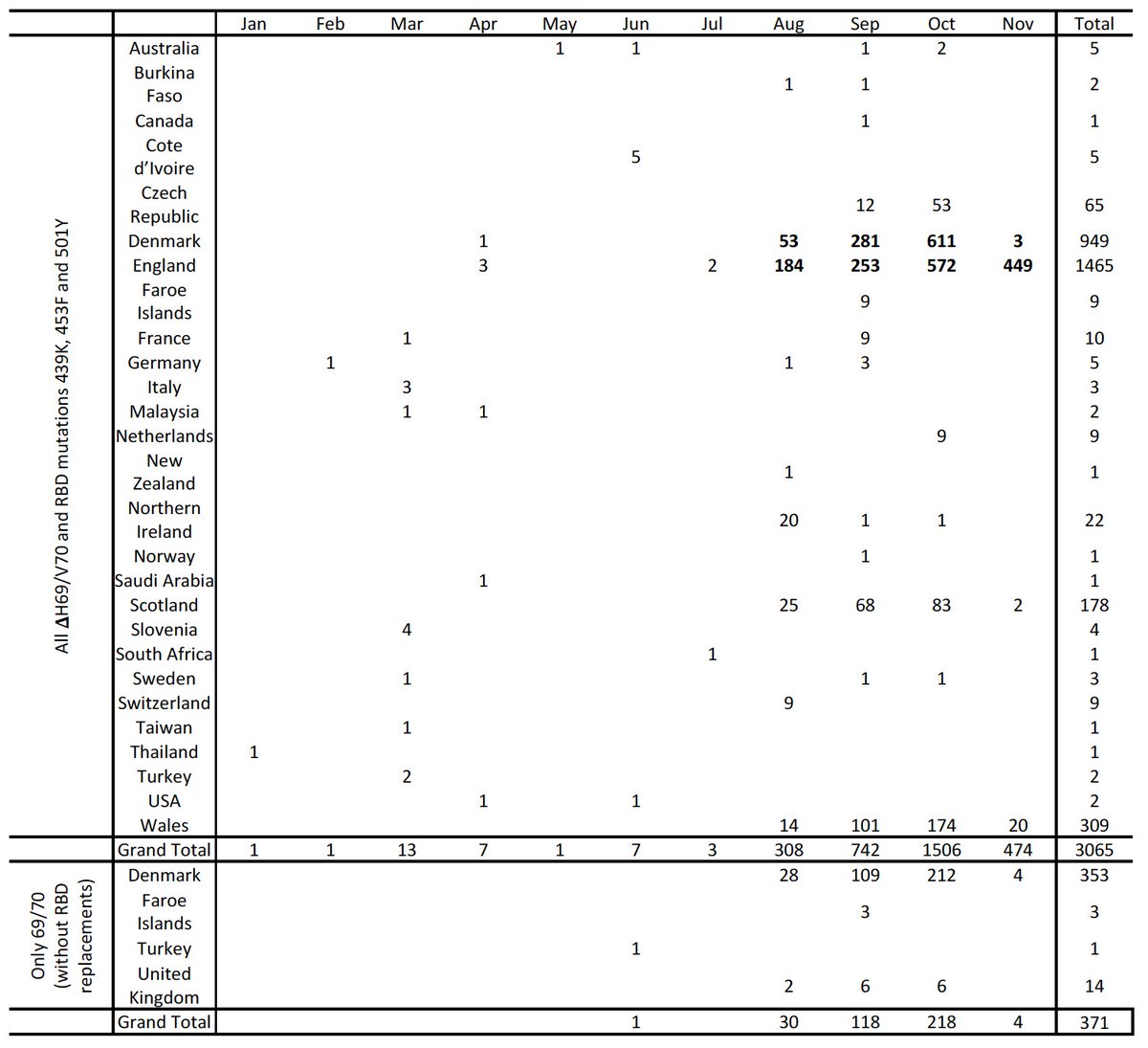
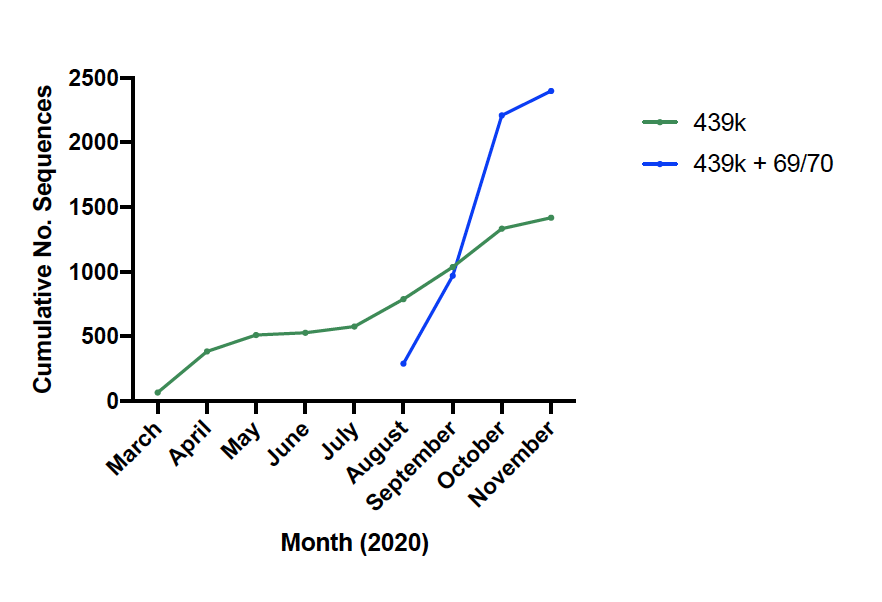
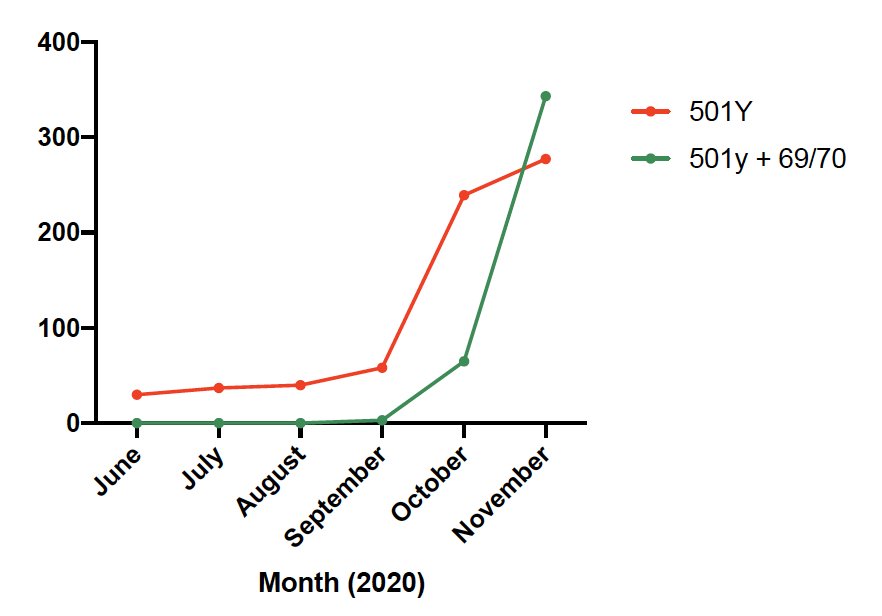
We identified an increase in the frequency of SARS-COV-2 sequences with the H69/V70 deletion, submitted to @GISAID- a global repository of SARS-COV-2 sequences. These variants started to expand in Denmark and the UK in August 2020.
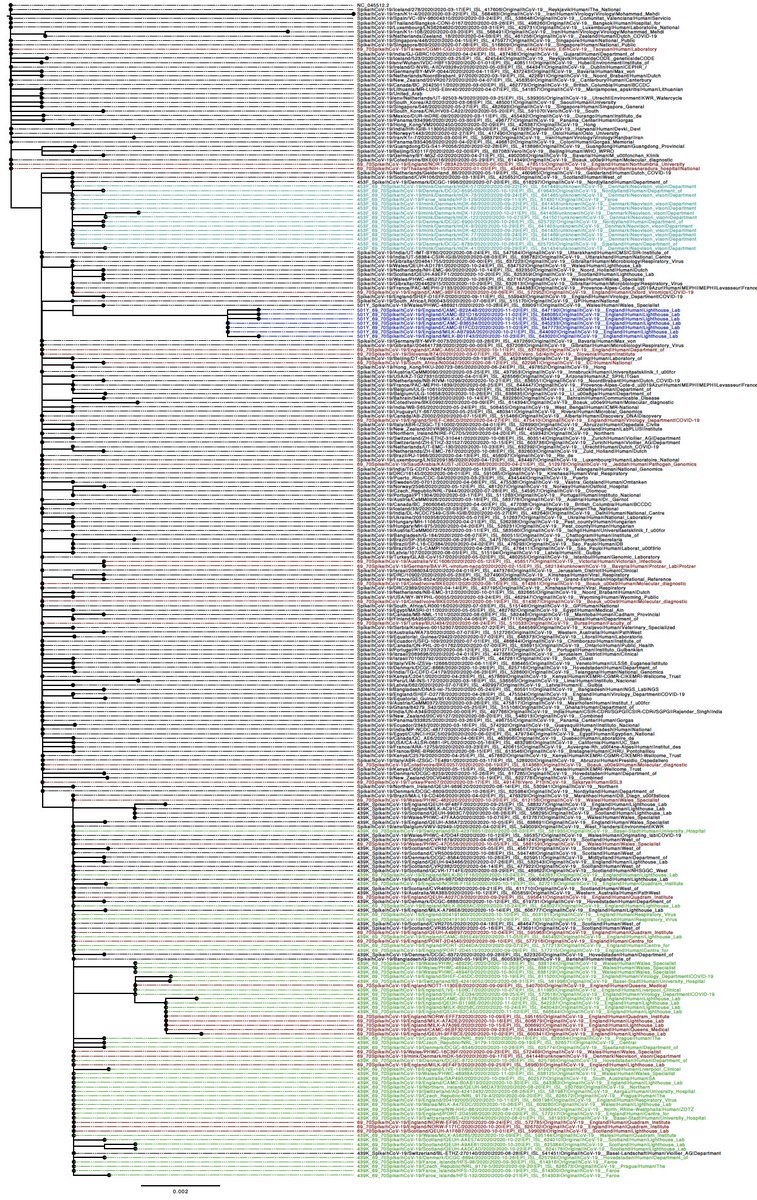
H69/V70 deletion occurs in many lineages globally. Either on their own (in red) or in concert with other mutations at the receptor binding domain; N501Y (dark blue), N453F (cyan) and Y439K (green).
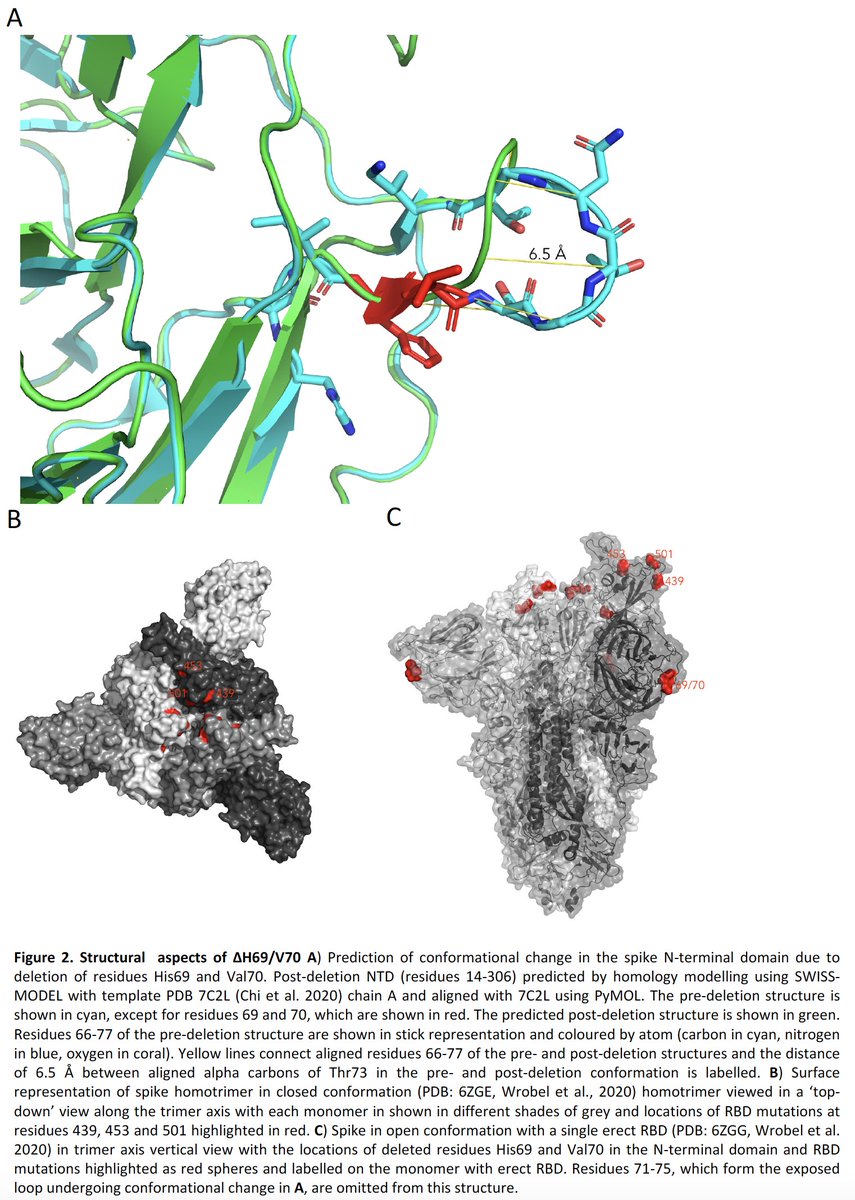
Structural analyses of the H69/V70 deletions in the NTD suggests a conformational change of the exposed loop by 6.5A, which we hypothesise may effect a conformational change in the RBD.
In addition to the above variants we discovered a distinct sub-lineage bearing the H69/V70 delations in addition to 6 linked Spike mutations (N501Y and A570D) in the RBD and (P681H,T716I, S982A and D1118H) in S2.
This is the variant described in the House of Commons today and is spreading rapidly in the South East of England and more widely. @ravgup33_ravi @CovidGenomicsUK @nhse @CMO_England @EricTopol @firefoxx66 @damicollier @rpdatir @steveo_kemp

 Read on Twitter
Read on Twitter