New preprint. We assessed the source of T-cell X-reactivity to #SARSCoV2 in unexposed people. Previous exposure to 'common cold' endemic HCoVs plays at most a marginal role, and we found no evidence for a yet-uncharacterised coronavirus in circulation.
1/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
1/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
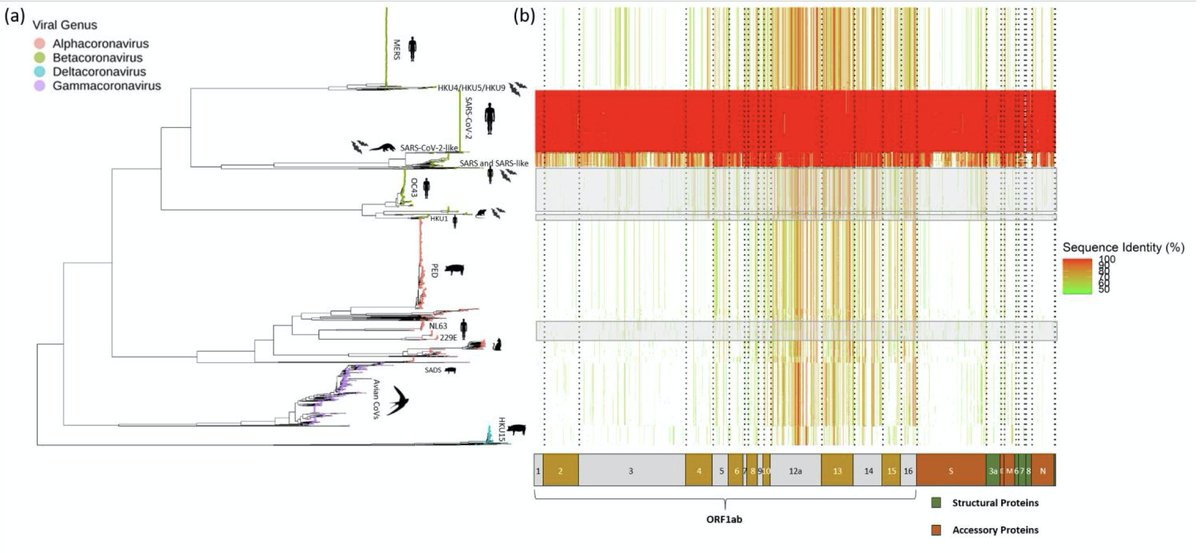
We built the most comprehensive phylogenetic tree for the 'orthocoronavirinae' to date and characterised all genomic regions with noticeable sequence homology to #SARSCoVV2 in any coronaviruses characterised at this stage.
1/
https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
1/
https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
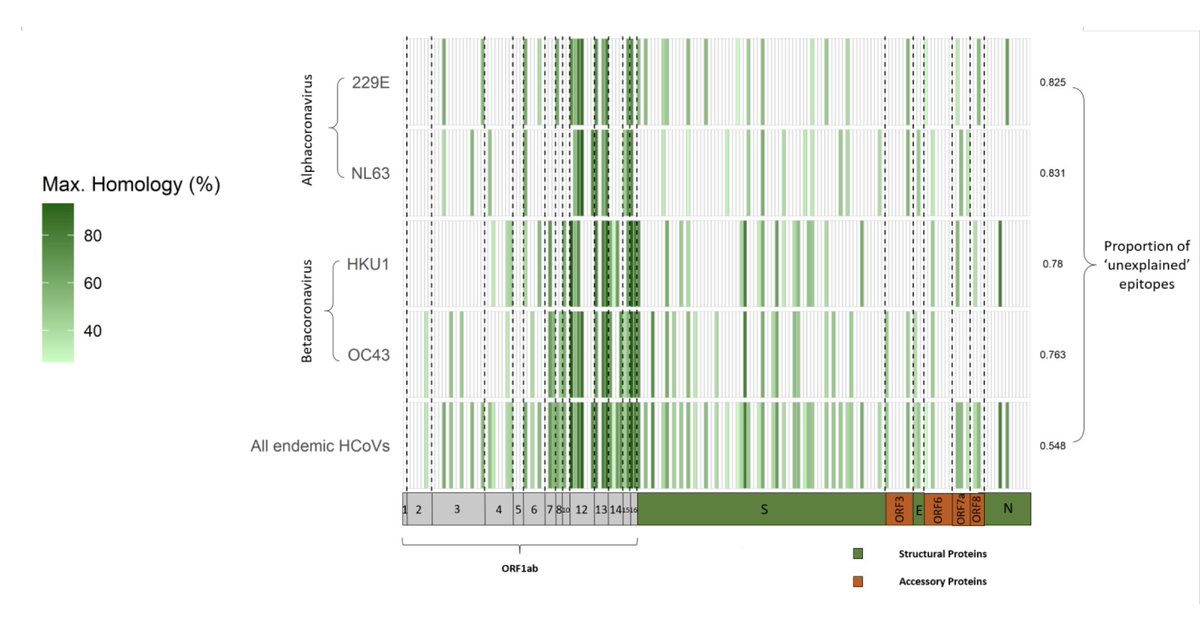
We then mapped onto the sequence alignment all 177 experimentally validated X-reactive CD4+/CD8+ epitopes reported to date in previously unexposed people. To our surprise, >50% of the epitopes have no noticeable homology with any of the 'common cold' HCoV.
https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
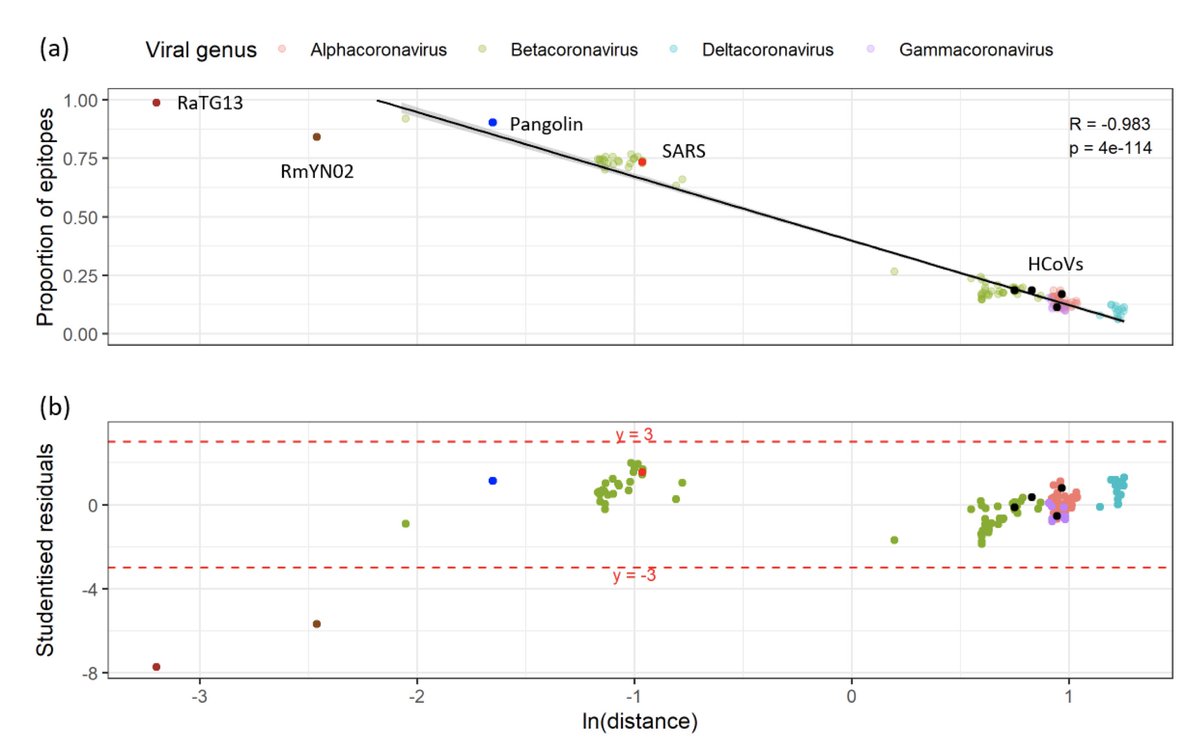
Moreover, even more surprisingly, none of the 155 coronavirus species known to date could explain a fraction of X-reactive epitopes above what would be expected simply by chance, once we accounted for their sequence identity with #SARSCoV2.
3/
https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
3/
https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
Taken together, our results indicate that the repertoire of X-reactive epitopes reported in healthy unexposed adults cannot be primarily explained by prior exposure to any coronavirus known to date, or any related yet-uncharacterised coronavirus.
4/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
4/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
Instead, patterns of pre-existing T-cell X-reactivity to #SARSCoV2 seem in line with lifelong exposure to a diverse and heterogenous array of various random antigens, with also no evidence for any vaccine-induced cross-reactivity (BCG or whatnot).
5/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
5/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
Our results don't directly inform on T-cell X-immunity, but they definitely do not support the idea of 'immunological dark matter', nor that prior exposure to HCoVs is a key factor in #SARSCoV2 susceptibility and/or symptom severity upon infection.
6/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
6/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
This has been a challenging project and the results don't fit with my preconceptions. Though, I'm grateful to all my collaborators for having been such wonderful companions. Thank you @CedricCSTan1, @ChrisJ_Owen, Christine, @Anto_Berto and @LucyvanDorp.
7/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1
7/ https://www.biorxiv.org/content/10.1101/2020.12.08.415703v1

 Read on Twitter
Read on Twitter