Our work to understand chromatin domains. Protein-mediated interactions hold chromatin segments together. How much can they fluctuate? How dynamic a domain is? Can they be super dynamic(liquid-like) or are they ~static(solid-like)? Our effort to estimate this: 1/6 @ranjithpa https://twitter.com/biorxivpreprint/status/1362338219476586498
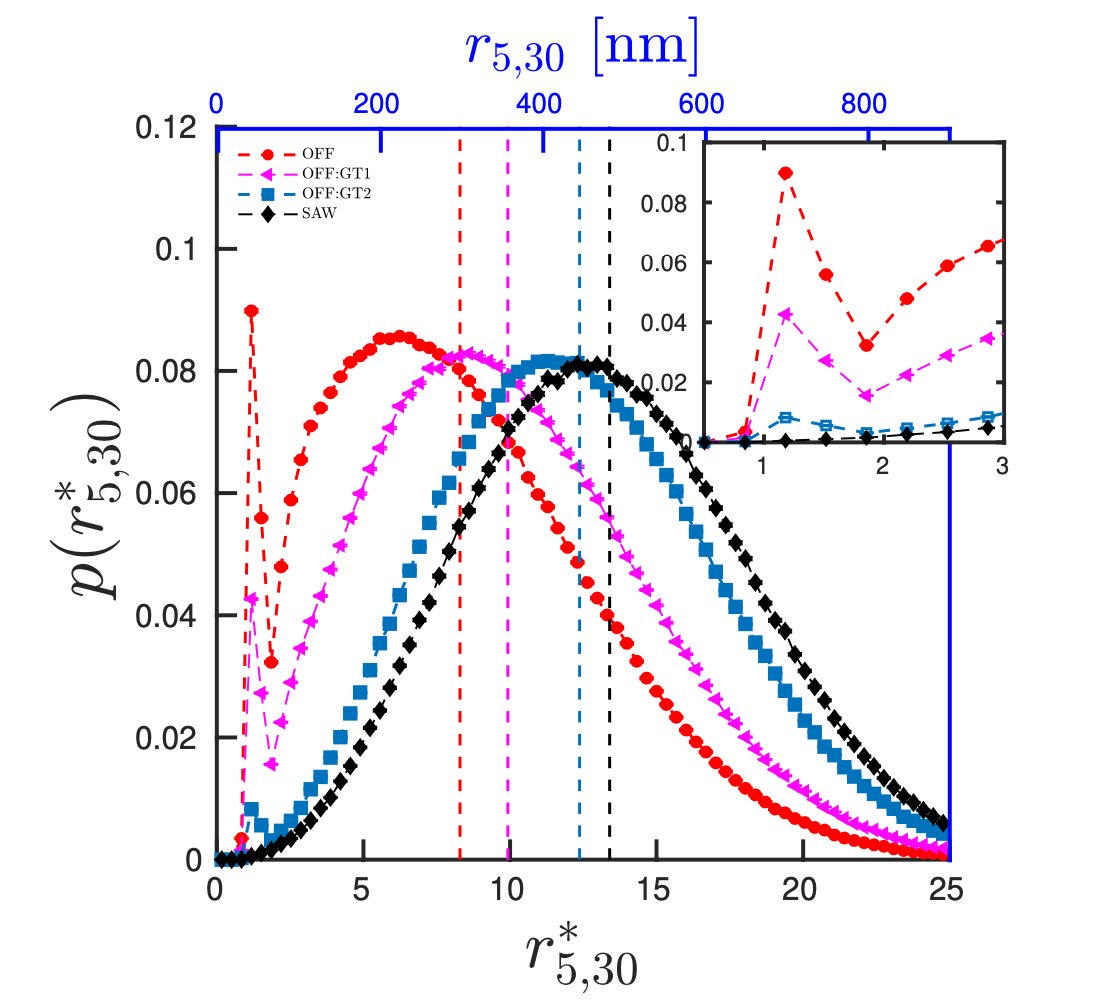
One of the challenges to simulate chromatin dynamics is that we do not know the interaction strengths (potential energies) between different segments of chromatin. We have developed a method that can estimate the interaction strengths, known the HiC-like contact maps. 2/6
We find: the average distances between segments cannot describe the complete picture of the domain. The distances have a wide distribution. Once the protein-mediated bond is broken, the polymer entropy can drive the segments far away. We have quantified this. 3/6
Meeting of two segments is assisted by co-operative folding of other regions. Interaction among segments can potentially influence meeting/fluctuation/folding of every other segment in the domain. 4/6
What is the typical volume of a domain? What is the shape of a domain? How much do they get altered as we vary interaction strengths/epigenetic states? We have quantified these. 5/6

 Read on Twitter
Read on Twitter