New work from the group published today, led by the incredible, integral, and incomparable @ruth_e_hanna, developing CRISPR Base Editor technology for screens.
Time for a hastily-assembled thread with screenshots! (1/n)….
Open access link here: https://www.sciencedirect.com/science/article/pii/S009286742100012X?dgcid=author
Time for a hastily-assembled thread with screenshots! (1/n)….
Open access link here: https://www.sciencedirect.com/science/article/pii/S009286742100012X?dgcid=author
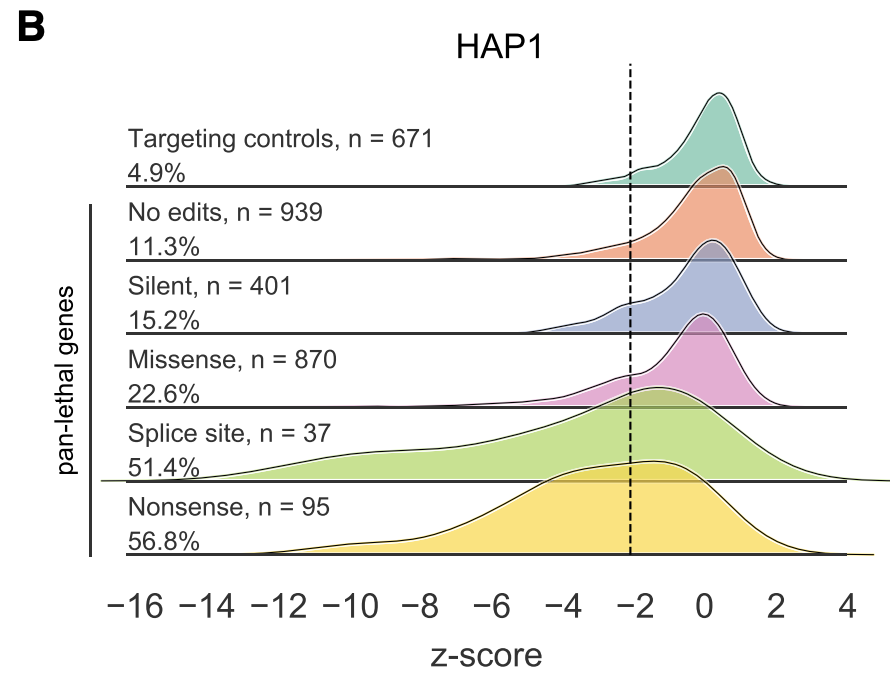
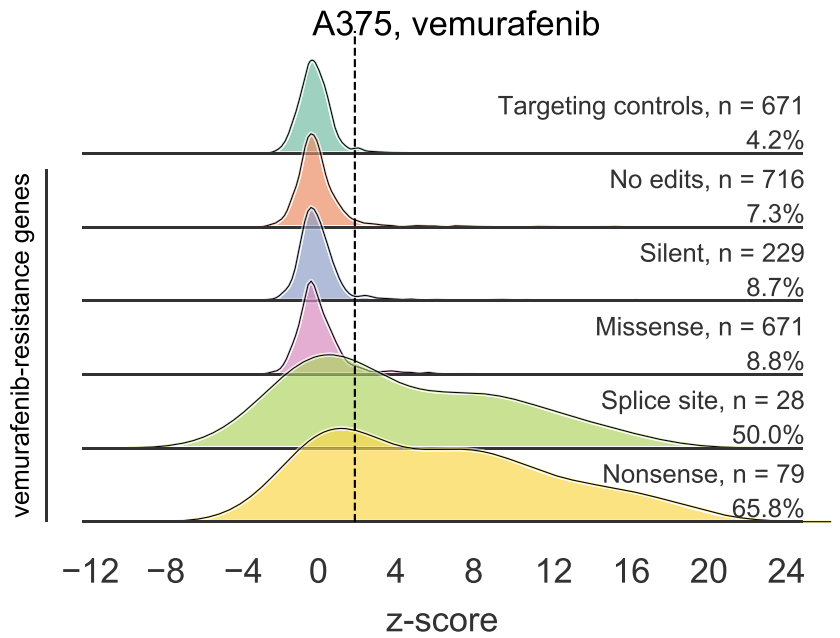
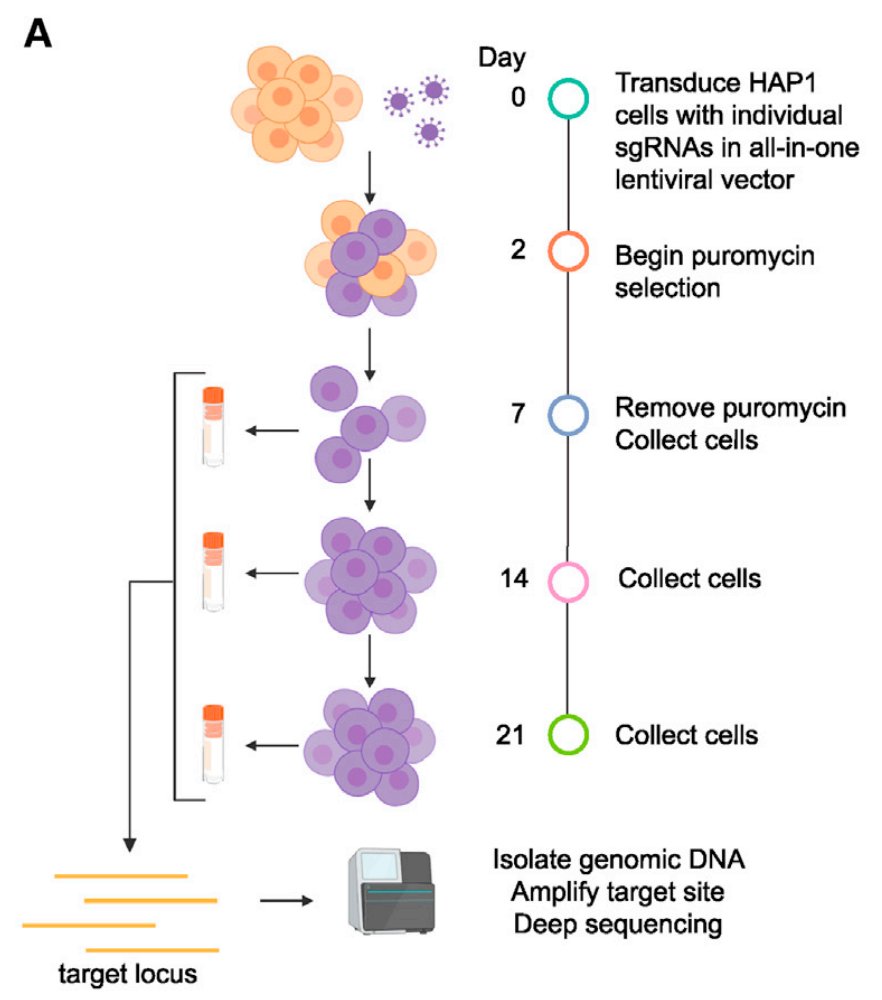
We made all-in-one lentiviruses to deliver a guide and the C>T Base editor (CBE). We performed negative selection screens (loss-of-function mutations in essential genes, assayed via growth) and positive selection (resistance to vemurafenib).
Two key takeaways from this:
1) Unexpected editing – outside of the intended window, creation of indels, etc. – is pretty rare. So that's good!
BUT...
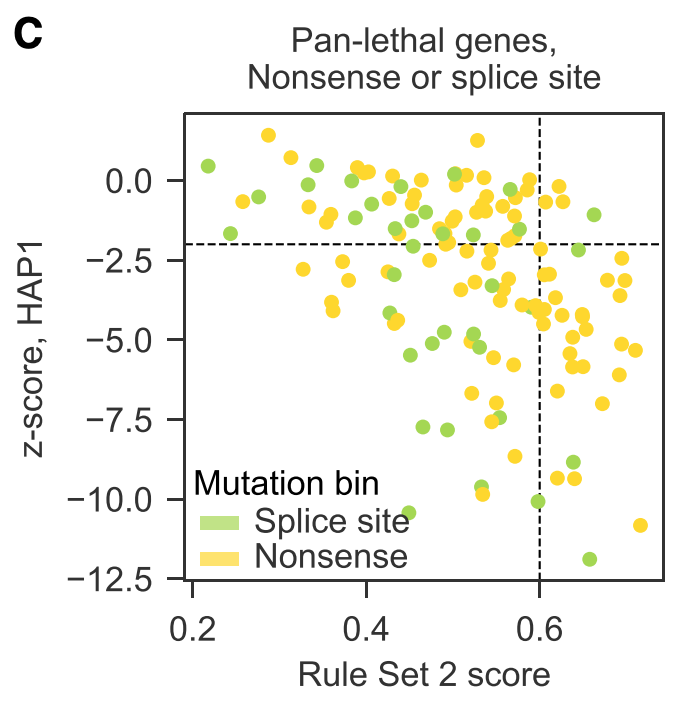
2) Certainly not the case that every guide works. Good ol' Rule Set 2 does a pretty good job of identifying effective guides.
1) Unexpected editing – outside of the intended window, creation of indels, etc. – is pretty rare. So that's good!
BUT...
2) Certainly not the case that every guide works. Good ol' Rule Set 2 does a pretty good job of identifying effective guides.
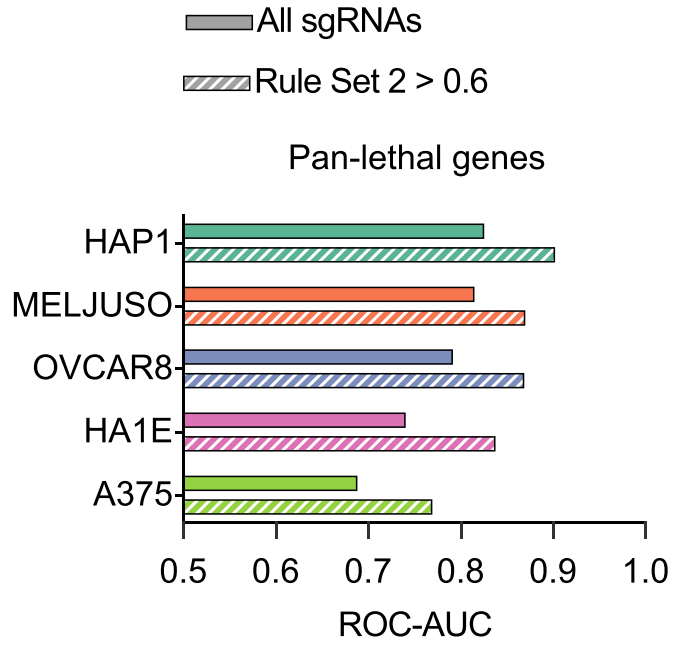
HAP1 cells being, generally, haploid, we also did negative selection screens in diploid or worse lines, and performance (higher value is better) reasonably consistent across cell lines
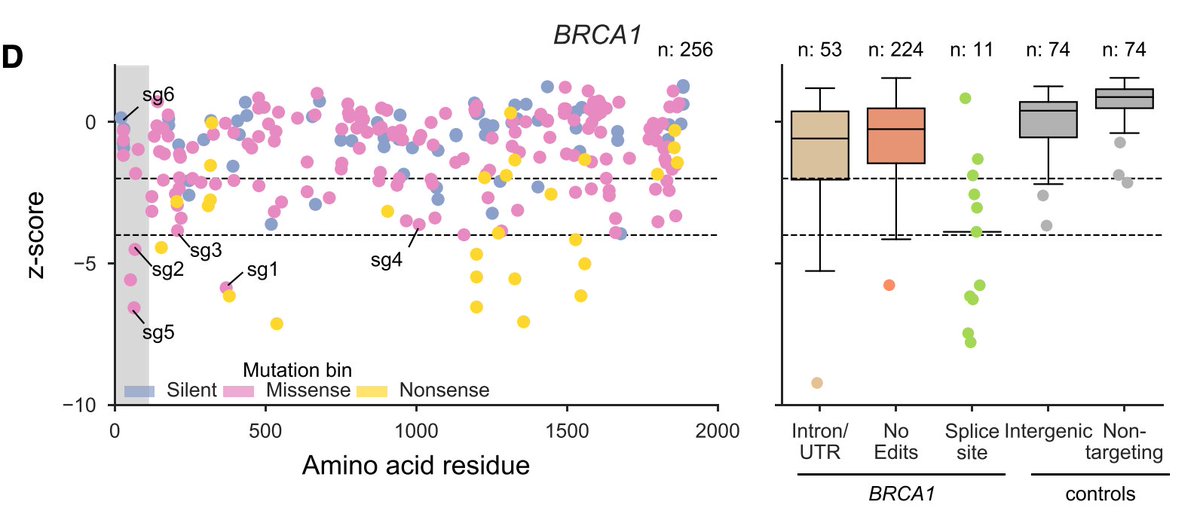
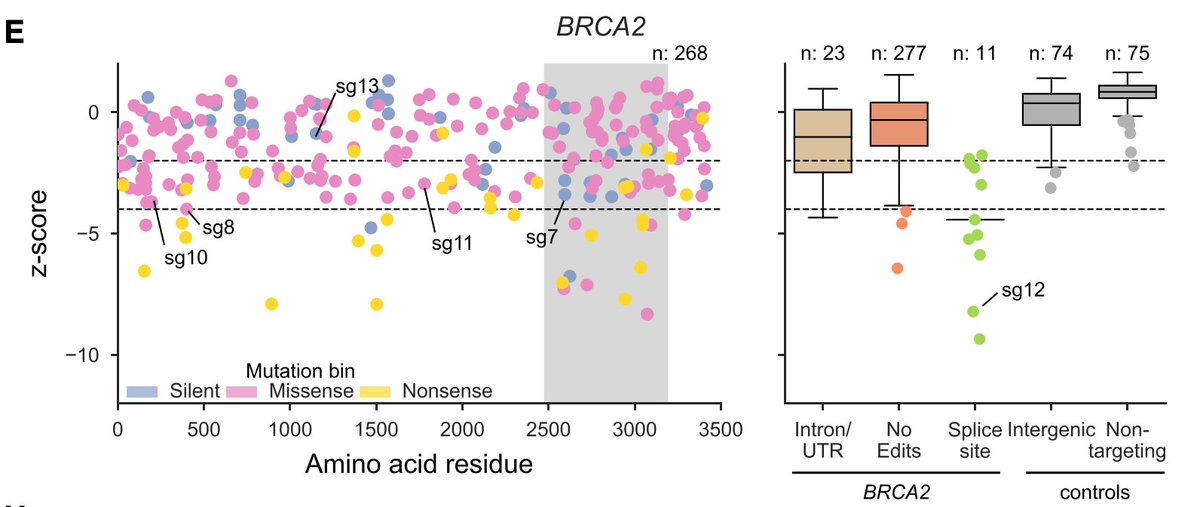
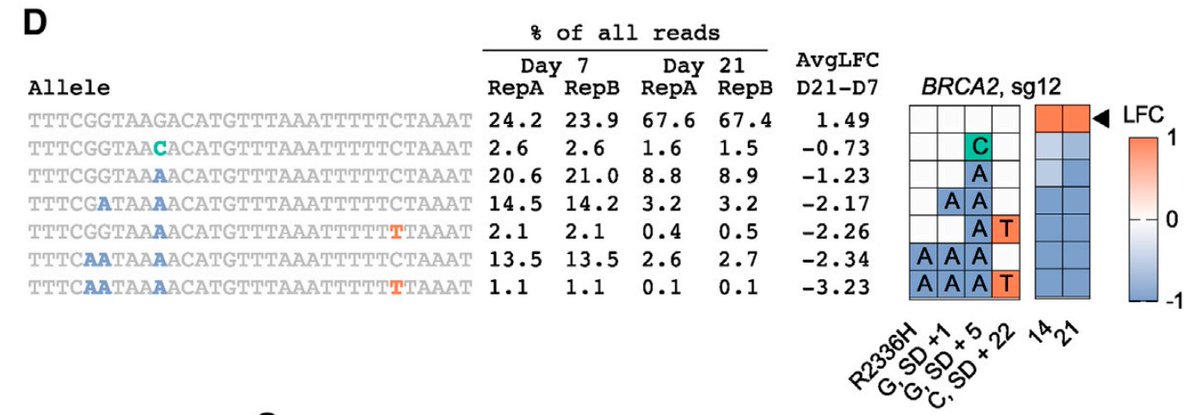
Next, we tiled across the genes BRCA1 and BRCA2, because there are an awful lot of Variants of Unknown Significance in ClinVar. Interestingly, most of the strongest missense mutations hit the RING domain (BRCA1) and DNA binding domain (BRCA2) (shaded region).
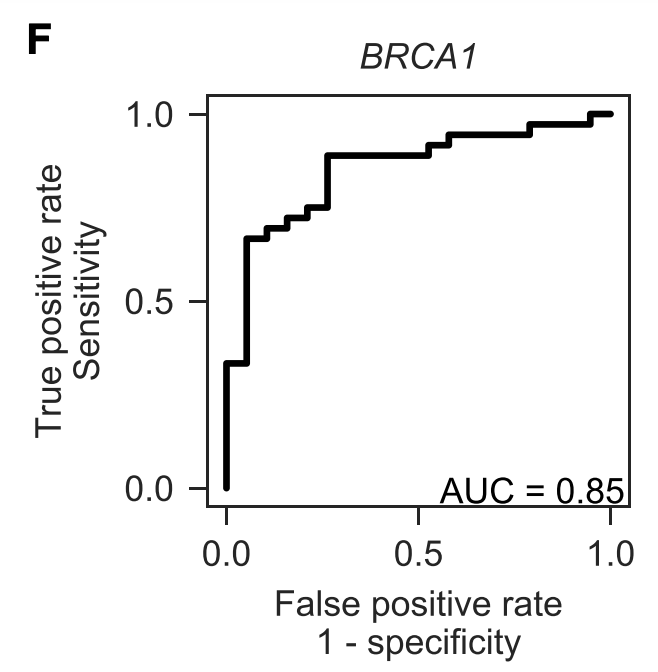
We can benchmark performance by assessing separation between variants classified as pathogenic or benign in ClinVar. Not perfect, and the sample size isn't huge, but still good to see.
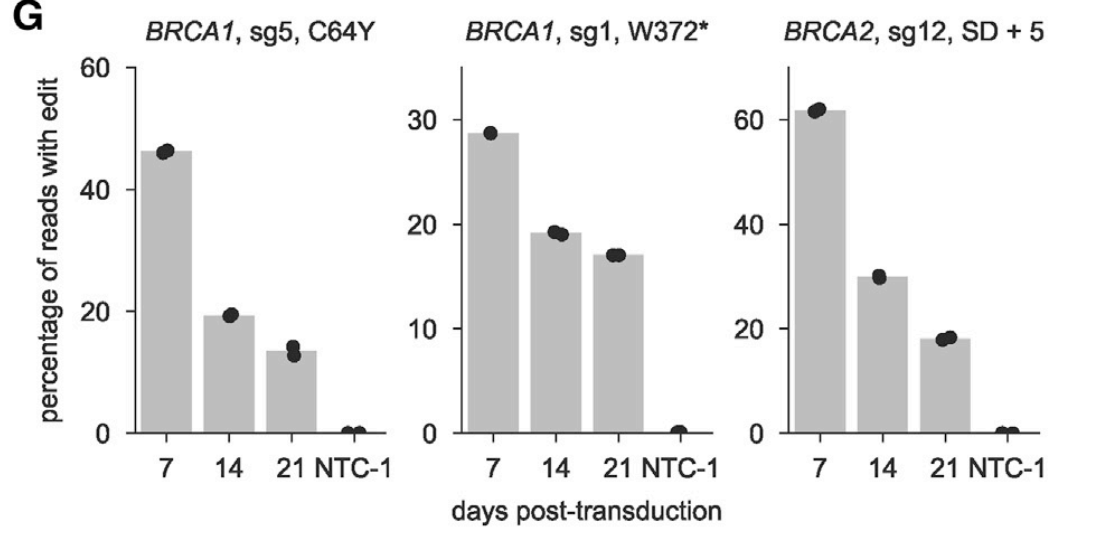
Up to this point, we're relying on the predicted base changes of the CBE, but need to validate. So for that, we introduce the guide one-at-a-time, PCR across the edit site, and sequence to see what actually happens at the genomic locus
Since this gives allele-level infomation, can figure out, in the case of multiple edits, driver vs. passenger. Here's a neat example of an intronic mutation that likely affects splicing – mutation at the +5 position necessary and sufficient for the decreased viability.
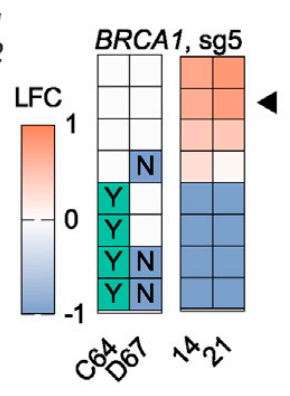
Here's another example, this time a protein-coding mutation, where C64Y drives the phenotype.
Another way to sum up this information is to group all reads by a mutation, can see that the C64Y mutation is selected against in the population over time.
Another way to sum up this information is to group all reads by a mutation, can see that the C64Y mutation is selected against in the population over time.
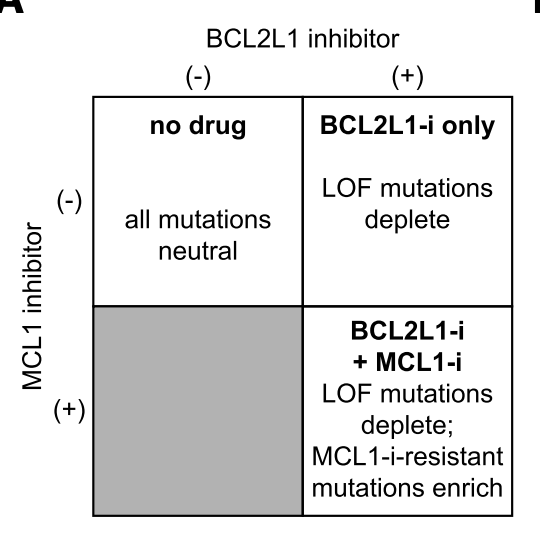
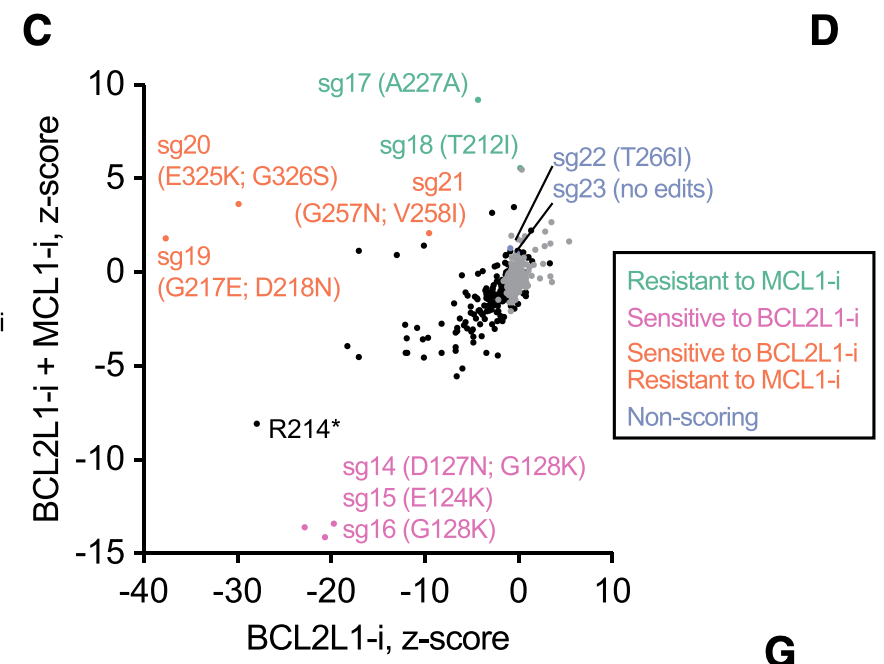
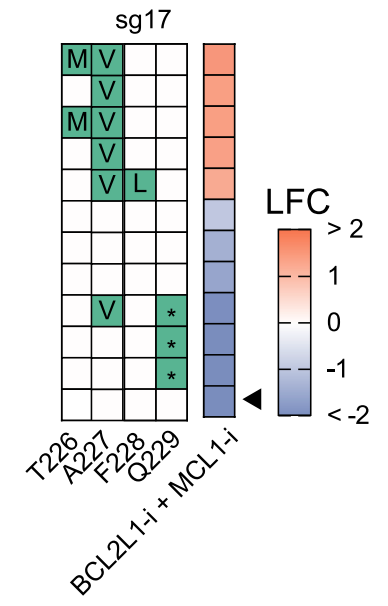
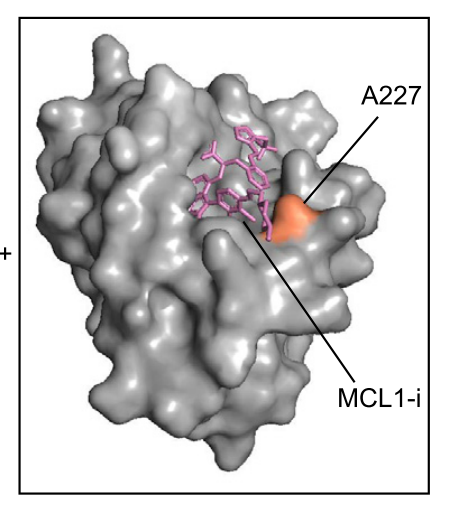
Turning our attention next to small molecule - protein interactions. Here, we could leverage the synthetic lethal relationship between MCL1 and BCL2L1 (aka BCL-xL) to find mutations that a) still maintain protein function but b) are resistant to small molecule inhibitors.
We found a lot of cool mutations that we are definitely not in the position to pursue further, so if you're an MCL1 or BCL2L1 afficianado, please let us know!
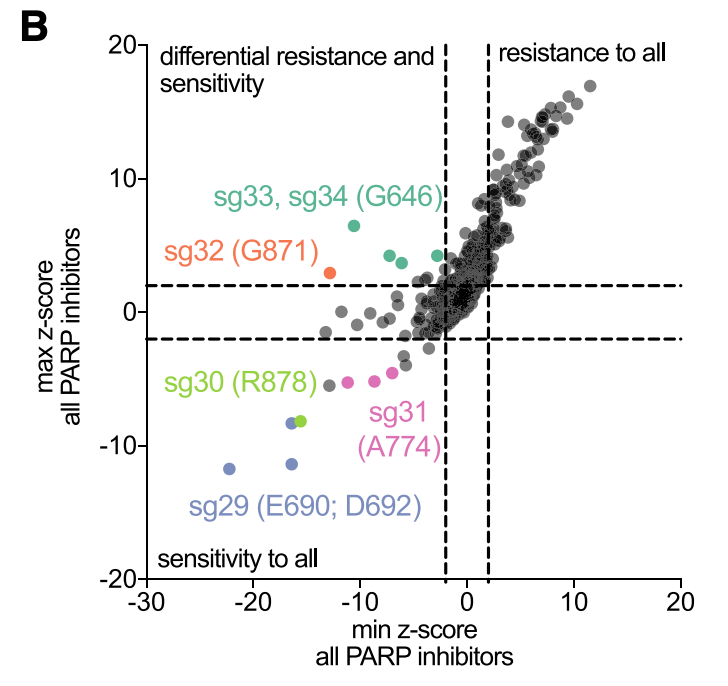
We took a similar approach for PARP inhibitors, and here we screened against five different small molecules in clinical use. Here, loss-of-function causes resistance (see work by @stveep). While most mutations affected all PARPi molecules the same, saw some differences emerge.
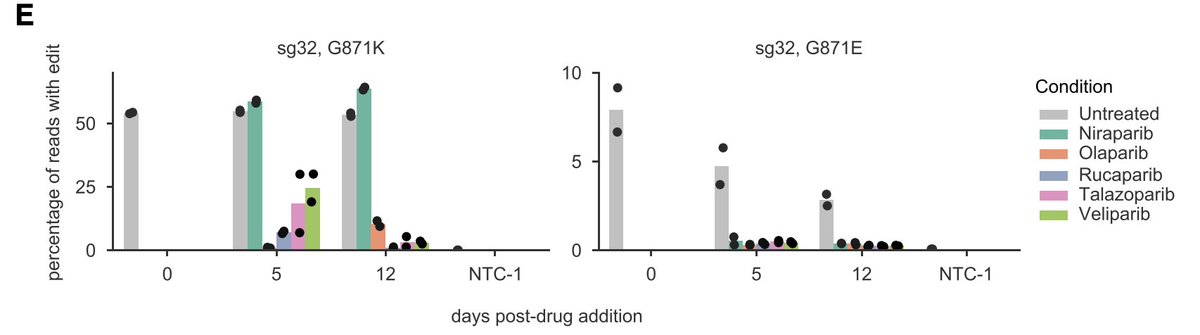
Here's one of my favorite examples. A G871K mutation sensitizes cells to 4 of the 5 PARP inhibitors, but not Niraparib. However, G871E mutation affects all 5 PARPi ~equally.
Getting this level of resolution can help guide additional rounds of small molecule optimization.
Getting this level of resolution can help guide additional rounds of small molecule optimization.
 SOAPBOX ALERT
SOAPBOX ALERT
I'd consider it Drug Development Malpractice™ to *not* do one of these screens. Identification of resistance mutations provides strong confidence that your small molecule is *actually* hitting the target you think it is.
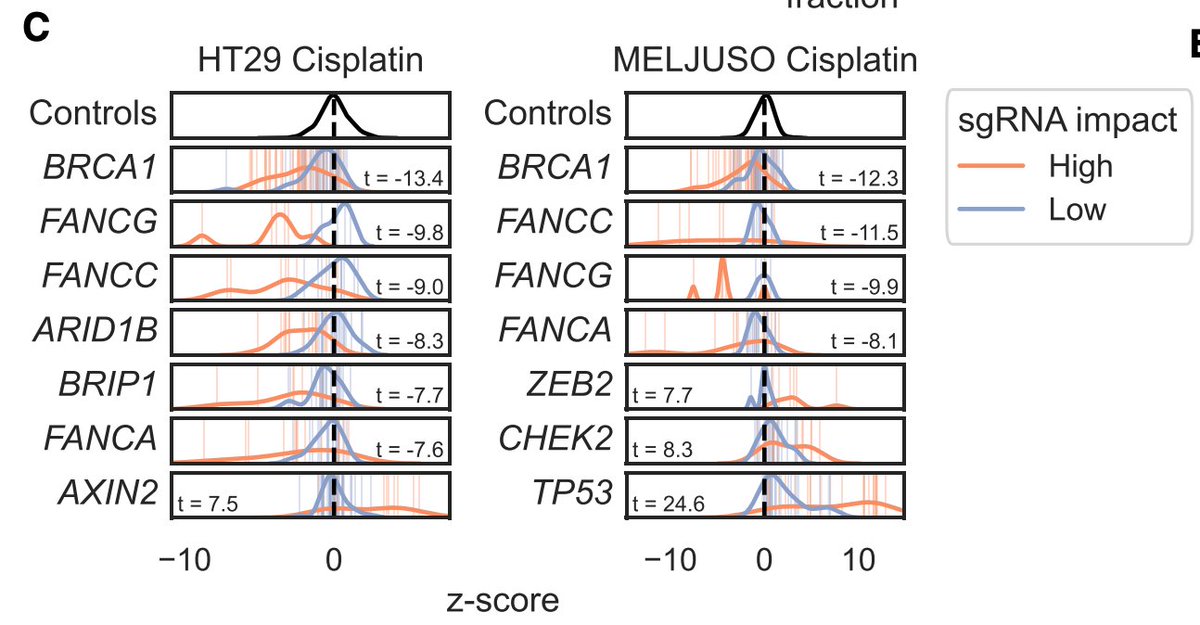
Okay, so how big can we go? We designed a library to introduce every C>T mutation in ClinVar, and screened for variants that modulated sensitivity to DNA damage (cisplatin) in two cell lines. Variants with a predicted high impact separated from low impact for many known genes.
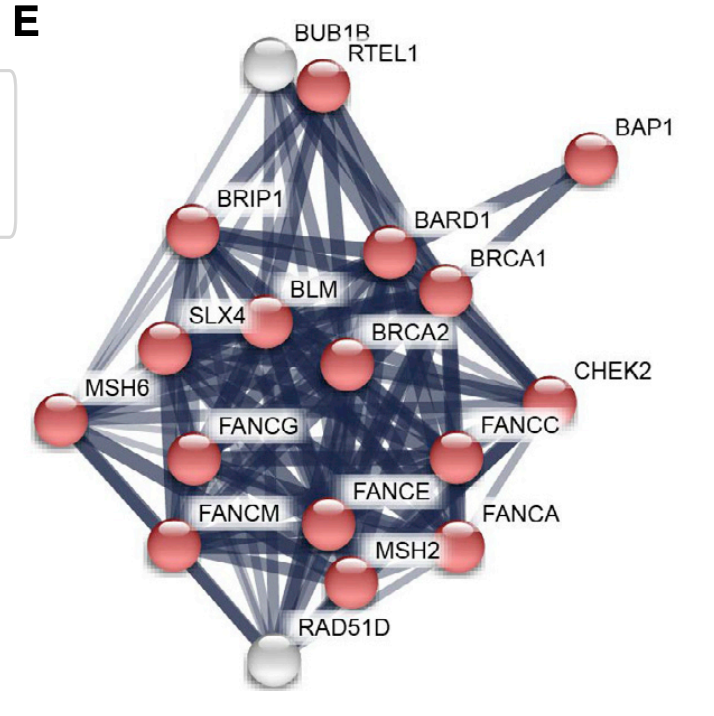
Genes that had many guides score are highly connected in String-db, as you'd expect.
If interested in DNA damage, also see a paper from @CicciaAlberto group!
If interested in DNA damage, also see a paper from @CicciaAlberto group!
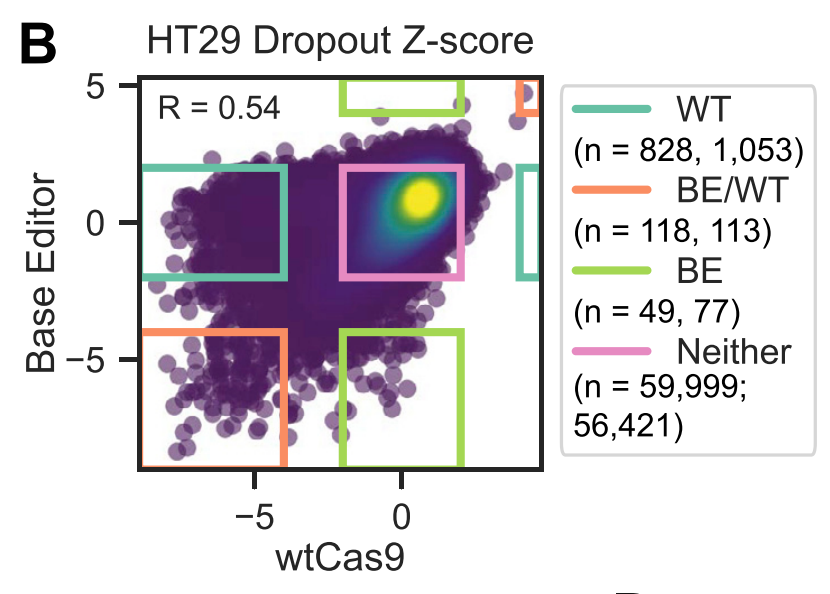
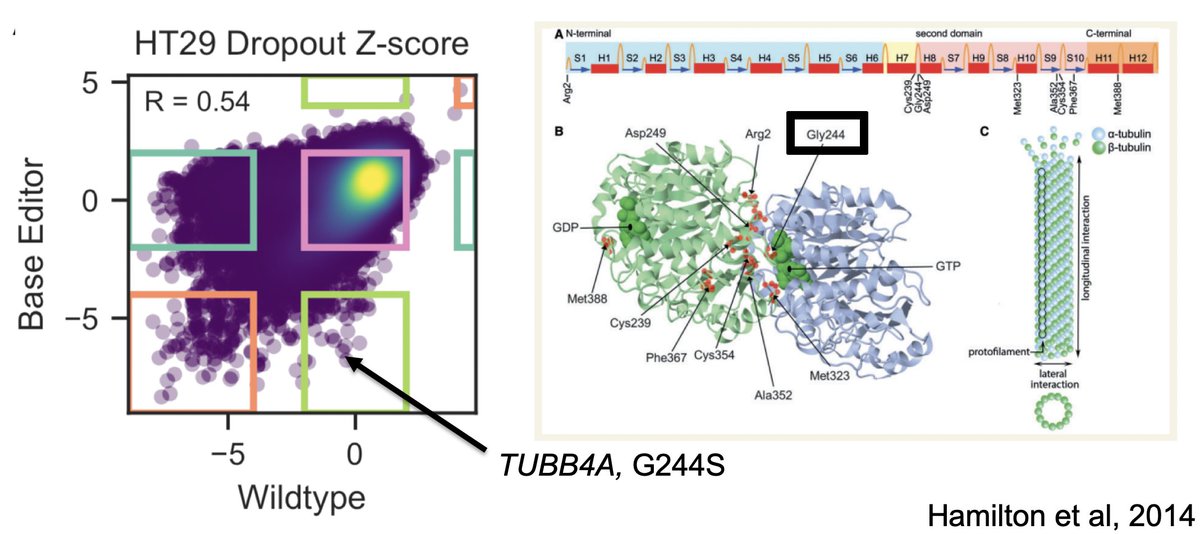
We also did a counter-screen with this library, using wt Cas9, i.e. for knockout. What's really cool about this is the identification of small number of guides that don't affect the cell with k/o, but *do* with a base edit, as possible gain-of-function mutations.
Here's one such example, a guide predicted to introduce G244S mutation in TUBB4A. When knocked out, other TUBB genes can compensate, but when this mutation is around / expressed, it prevents proper assembly of tubulin assembly. Pretty nifty!
In addition to the now-departed Ruth Hanna* thanks to fellow tweeting GPPers @HegdeMudra @p_deweirdt @MarissaFeeley as well as collaborators @jamesTneal @davidrliu @LukeKoblan
*to @MITBiology for grad school!
Happy to take questions!
*to @MITBiology for grad school!
Happy to take questions!

 Read on Twitter
Read on Twitter