In a new @biorxivpreprint with @McLellan_Lab @ltchong @Terrasztain @LCasalino88 @fiona_chembot @JoryGoldsmith, we show how a glycan gate (at position N343) controls the opening of the #SARSCoV2 spike protein
#GlycanGate #glycotime
https://www.biorxiv.org/content/10.1101/2021.02.15.431212v1
 :
:
we use > 200 microseconds weighted ensemble (WE) MD simulations to generate > 300 continuous, kinetically unbiased spike opening pathways
& in the process set a new high-water mark for WE simulations
@TACC @WestpaSoftware
& in the process set a new high-water mark for WE simulations

@TACC @WestpaSoftware
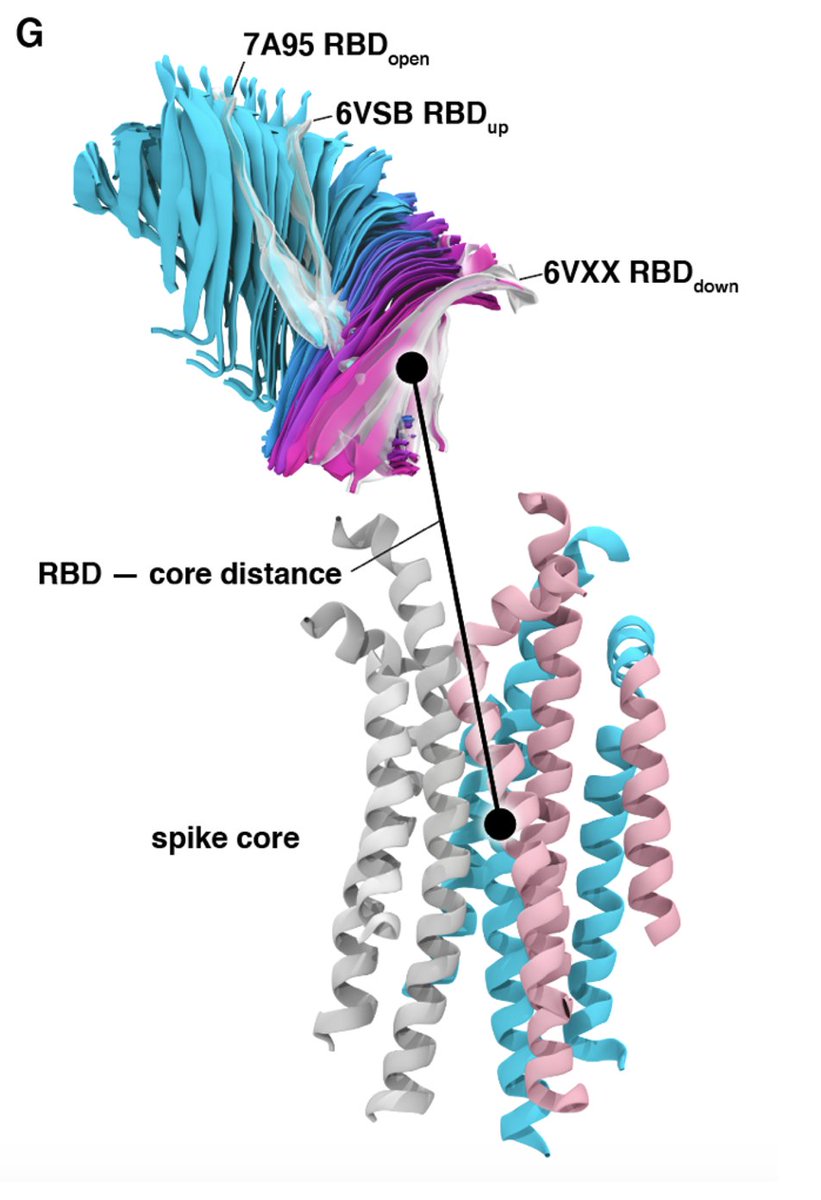
Analysis of these pathways & experiments reveal R408, D405, D427 participate in the RBD opening mechanism & the N343 glycan acts as a “gate” – initially blocking but then pushing up the RBD by intercalating between residues F490, Y489, F456, and R457 of the ACE2 binding motif
and yes, it did also look to us like the glycan at N343 was doing the “hand-jive” with the other residues there!
The RBD in our simulations continues to open, spontaneously, beyond the ‘up’ state in 6VSB, beyond the ‘open’ state in 7A95, to an even more widely open state en route to S1 disassociation (similar to what @drGregBowman @ElisaTelisa @foldingathome team found)
BTW none of the emerging mutant #SARSCoV2 strains that we’ve looked at, including #UKvariant #B1117 #BrazilianVariant #P1 #SouthAfricanVariant #B1351 and CAL.20C, contain mutations in the residues we identified here to facilitate RBD opening. 

Example opening trajectory & system files available at http://amarolab.ucsd.edu
& thank you to @TACC @NSF @NIH @RCSA for compute time & support
& thank you to @TACC @NSF @NIH @RCSA for compute time & support
& yes - I really did delete the original tweet and redo this whole thing because even though this is @Twitter- I'm an academic and #ProperCreditMatters 
\\end

\\end

 Read on Twitter
Read on Twitter