About a year ago we shared this picture of 40yr old phage stocks and asked "what should we do with them?" 
Replies: 'check titre!' 'get WGS' and 'are they the same as the stock phages we've all been using?'
Well.. today out in @mbio a paper by @Dinessubedi did just this! https://twitter.com/JeremyJBarr/status/1157060823837057025

Replies: 'check titre!' 'get WGS' and 'are they the same as the stock phages we've all been using?'
Well.. today out in @mbio a paper by @Dinessubedi did just this! https://twitter.com/JeremyJBarr/status/1157060823837057025
Our paper compared the stability, infectivity and genetics of these 48yr old phages (shout out to Bob @thehancocklab who isolated these phages during his PhD!) to contemporary phages from two labs - @thebarrlab_aus and Rao lab https://www.t4lab.net/ https://msystems.asm.org/content/6/1/e00990-20
So what did we find?
- From the T2, T3, T4, T6 & T7 phages, only the T-even phages were recoverable - potential long-term stability of T-even capsid structure?
- Despite nearly 50years in solitary isolation, T2 & T6 phage genomes only had 12 & 16 SNPs compared to NCBI genomes
- From the T2, T3, T4, T6 & T7 phages, only the T-even phages were recoverable - potential long-term stability of T-even capsid structure?
- Despite nearly 50years in solitary isolation, T2 & T6 phage genomes only had 12 & 16 SNPs compared to NCBI genomes
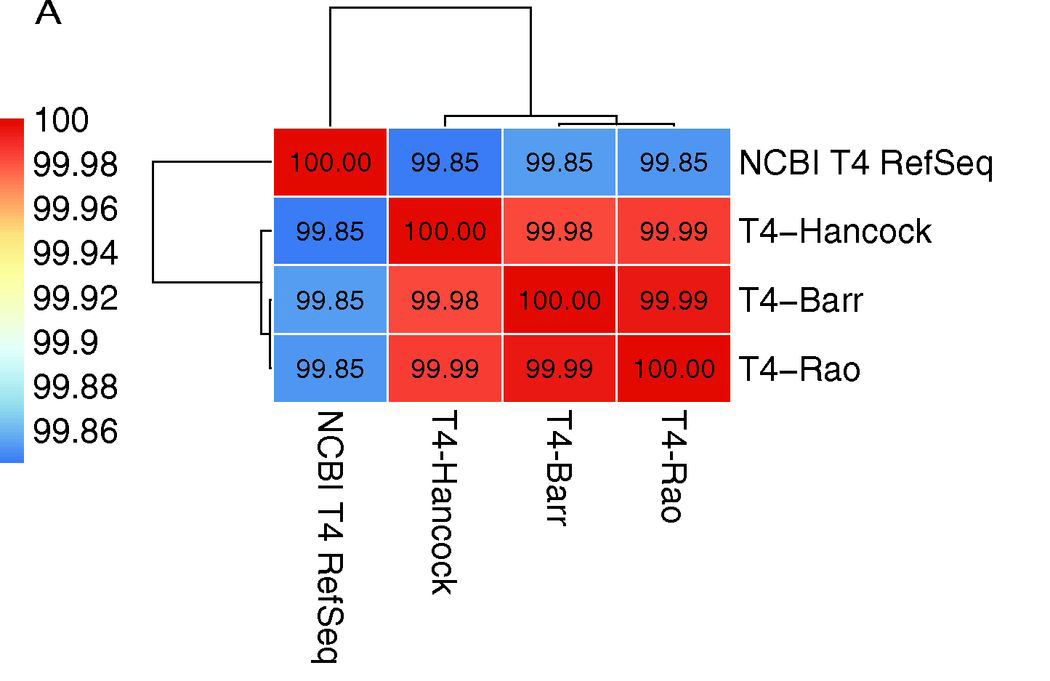
T4 phage however, had a staggering 172 nucleotide variations compared to NCBI ref!
We WGS the 48yr old phage (Hancock) and two active T4 phages from Barr & Rao labs and found between 6-13 variations between them.
The issue? T4 NCBI ref genome was assembled via PCR & cloning
We WGS the 48yr old phage (Hancock) and two active T4 phages from Barr & Rao labs and found between 6-13 variations between them.
The issue? T4 NCBI ref genome was assembled via PCR & cloning
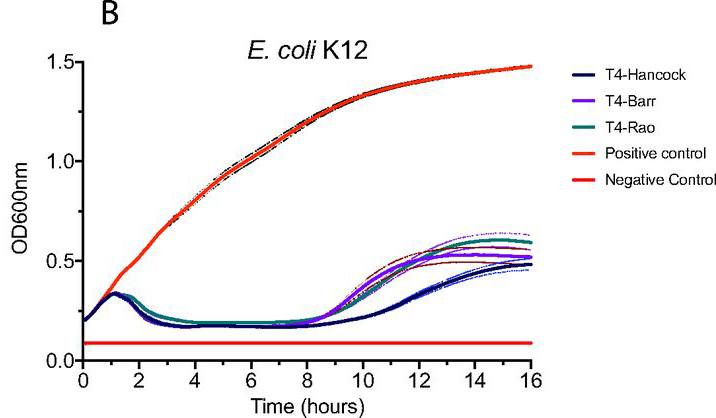
Finally we compared in vitro activity of all the T4 phages - and despite being on ice for nearly 50 years and ~10 variations in auxiliary & essential genes - we found no differences in adsorption, EOP, latent period, burst size or antimicrobial effect against E.coli B or K12
Our take home points.
- These phage stocks are incredibly value, we're in process of depositing them to NTCC (& happy to share!)
- Genetic & phenotypic drift of T-even phage across 48yrs was minor
- Delbruck & co's rationale for the use of comparable model phages still holds true
- These phage stocks are incredibly value, we're in process of depositing them to NTCC (& happy to share!)
- Genetic & phenotypic drift of T-even phage across 48yrs was minor
- Delbruck & co's rationale for the use of comparable model phages still holds true
Paper out in @mSystemsJ ! best to tag the correct journal 


 Read on Twitter
Read on Twitter