Where might novel coronaviruses come from? Our study with @Baylism and Marcus Blagrove is now out:
https://www.nature.com/articles/s41467-021-21034-5
@LivUni_IVES
Thread 1/16
https://www.nature.com/articles/s41467-021-21034-5
@LivUni_IVES
Thread 1/16
Background:
Novel coronaviruses arise by homologous recombination of two different coronavirus strains co-infecting an animal host.
These novel viruses circulate in animal populations before spillover to humans and may exploit liaison hosts in this multi-host spread.
2/16
Novel coronaviruses arise by homologous recombination of two different coronavirus strains co-infecting an animal host.
These novel viruses circulate in animal populations before spillover to humans and may exploit liaison hosts in this multi-host spread.
2/16
homologous recombination?
If an animal is infected with two coronaviruses at the same time, these viruses can ‘recombine’ – or swap – parts of their genetic material.
This can result in a daughter virus being generated with parts from each of the two parent viruses.
3/16
If an animal is infected with two coronaviruses at the same time, these viruses can ‘recombine’ – or swap – parts of their genetic material.
This can result in a daughter virus being generated with parts from each of the two parent viruses.
3/16
homologous recombination?
For example, the daughter virus may ‘inherit’ genes from one parent which allow it to infect specific animals, and genes from the other parent which cause specific disease symptoms.
This new virus has a unique combination of genes.
4/16
For example, the daughter virus may ‘inherit’ genes from one parent which allow it to infect specific animals, and genes from the other parent which cause specific disease symptoms.
This new virus has a unique combination of genes.
4/16
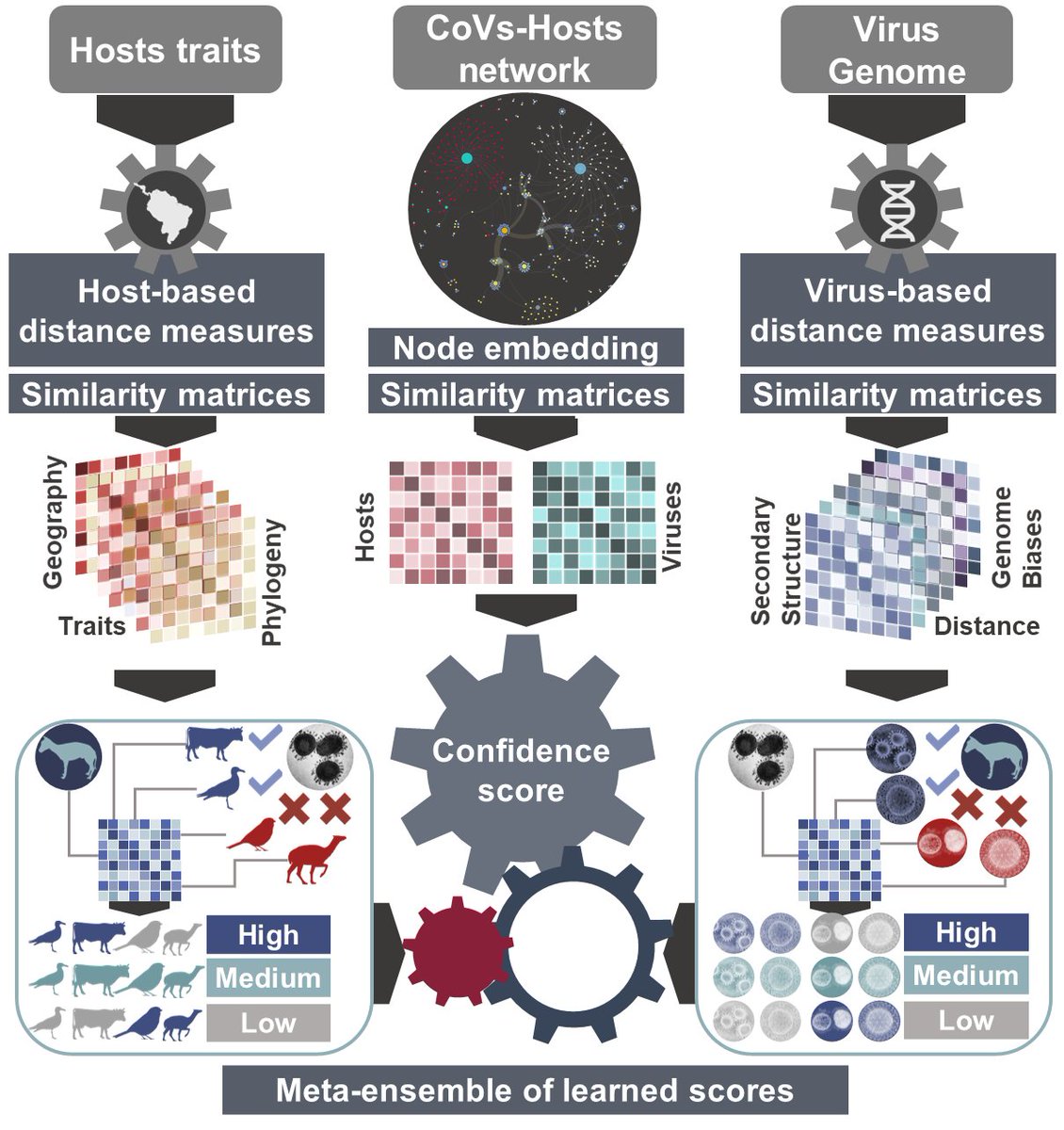
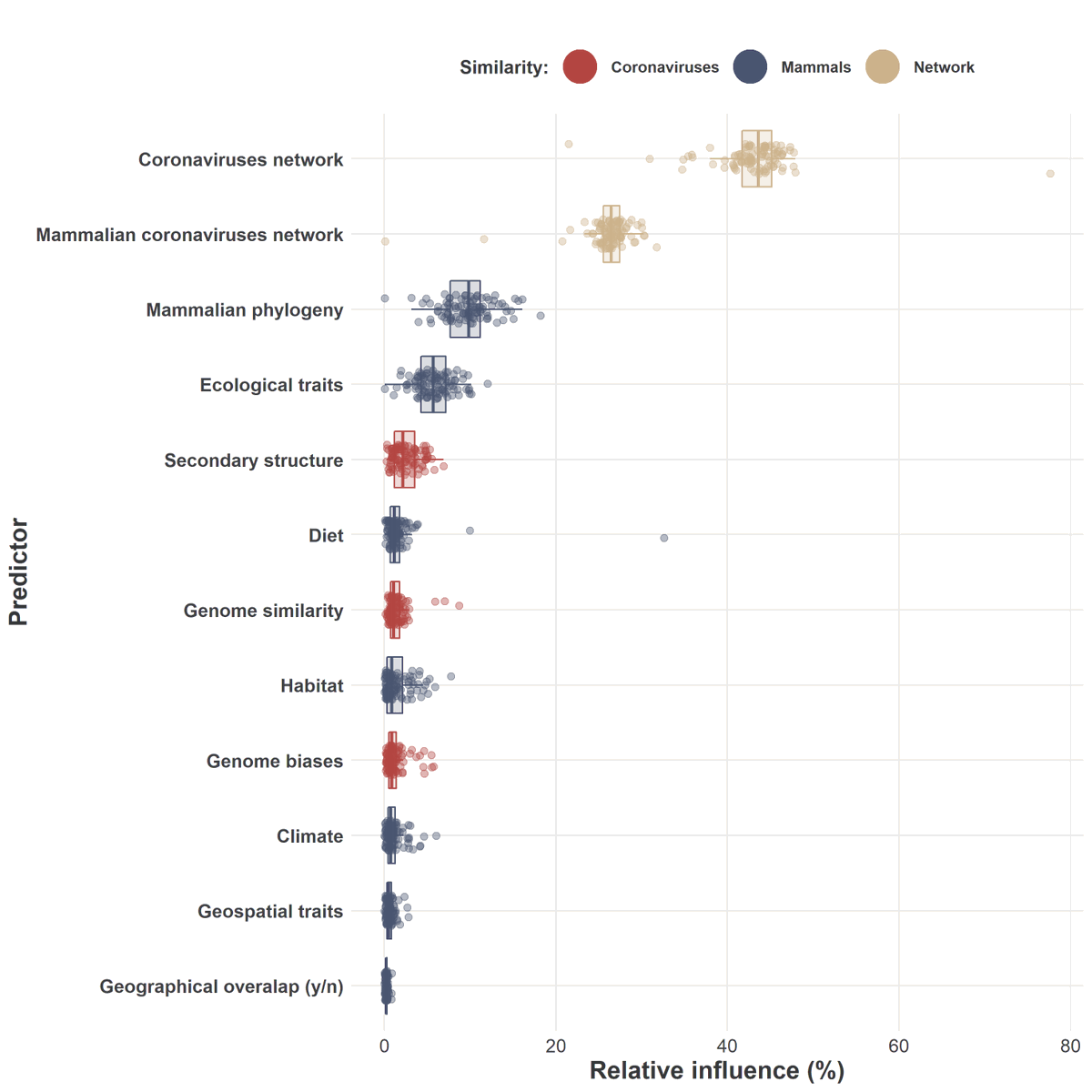
How did we do it?
This problem is a many-sided puzzle.
We identified key factors from the mammal side such as: its phylogenetic distance to know hosts of each coronavirus, diet, and the type of habitat it lives in, also in relation to known hosts.
5/16
This problem is a many-sided puzzle.
We identified key factors from the mammal side such as: its phylogenetic distance to know hosts of each coronavirus, diet, and the type of habitat it lives in, also in relation to known hosts.
5/16
How did we do it?
From the virus side we used: the genome sequence, the ‘secondary structure’ – or physical shape – of the genome, and the ‘biases’ – or frequency – of combinations of ‘bases’ – or letters – in the genome.
6/16
From the virus side we used: the genome sequence, the ‘secondary structure’ – or physical shape – of the genome, and the ‘biases’ – or frequency – of combinations of ‘bases’ – or letters – in the genome.
6/16
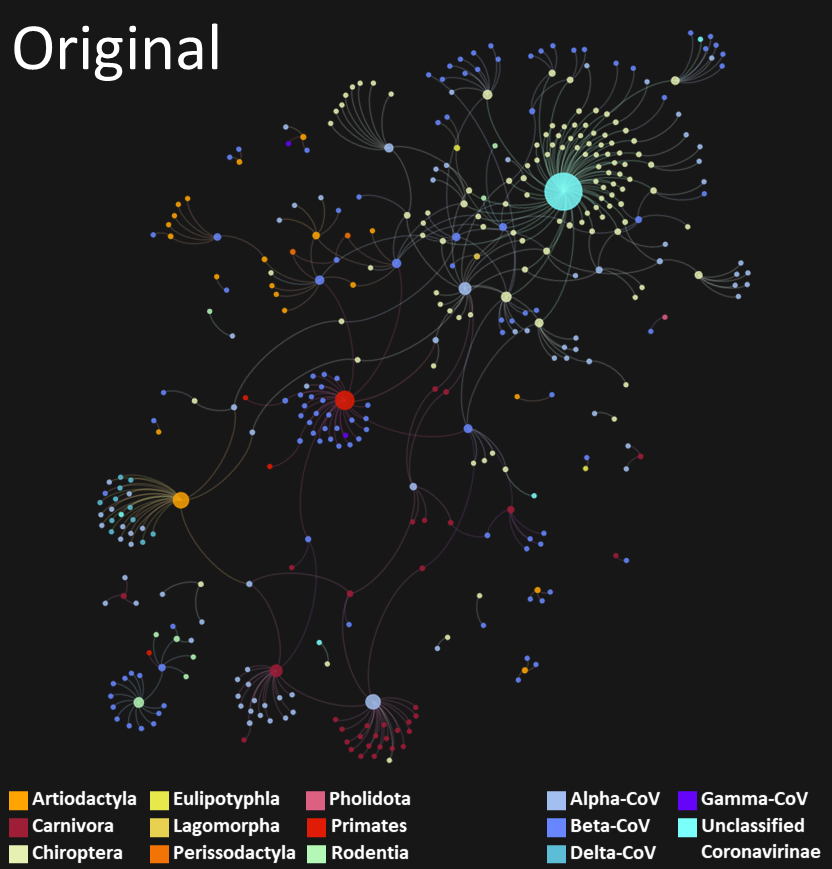
How did we do it?
Finally, we also used the network linking known coronaviruses and their known hosts, and this enabled us to find the last side of the puzzle: the complex connections that already exist between coronaviruses and mammals.
7/16
Finally, we also used the network linking known coronaviruses and their known hosts, and this enabled us to find the last side of the puzzle: the complex connections that already exist between coronaviruses and mammals.
7/16
How did we do it?
we then blended all these different sides together - we built an ensemble using GBMs (we compared many other stacking algoritms, GBMs was the most reliable).
8/16
we then blended all these different sides together - we built an ensemble using GBMs (we compared many other stacking algoritms, GBMs was the most reliable).
8/16
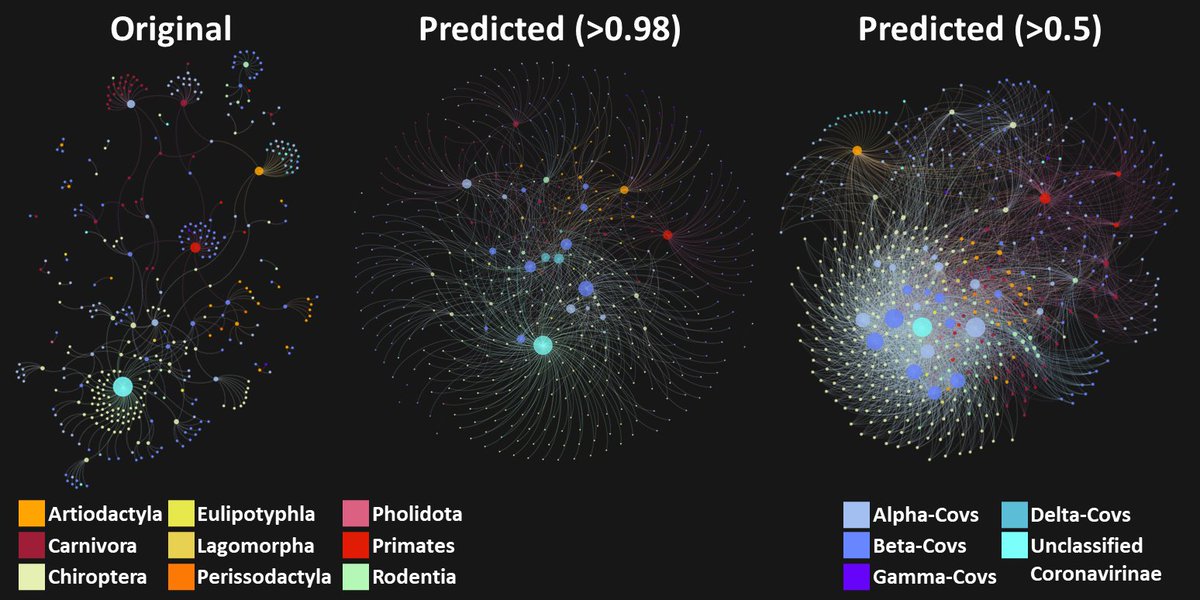
Our results:
We predict that there are at least 11 times more associations between mammalian species and coronavirus strains than have been observed to date.
9/16
We predict that there are at least 11 times more associations between mammalian species and coronavirus strains than have been observed to date.
9/16
Our results:
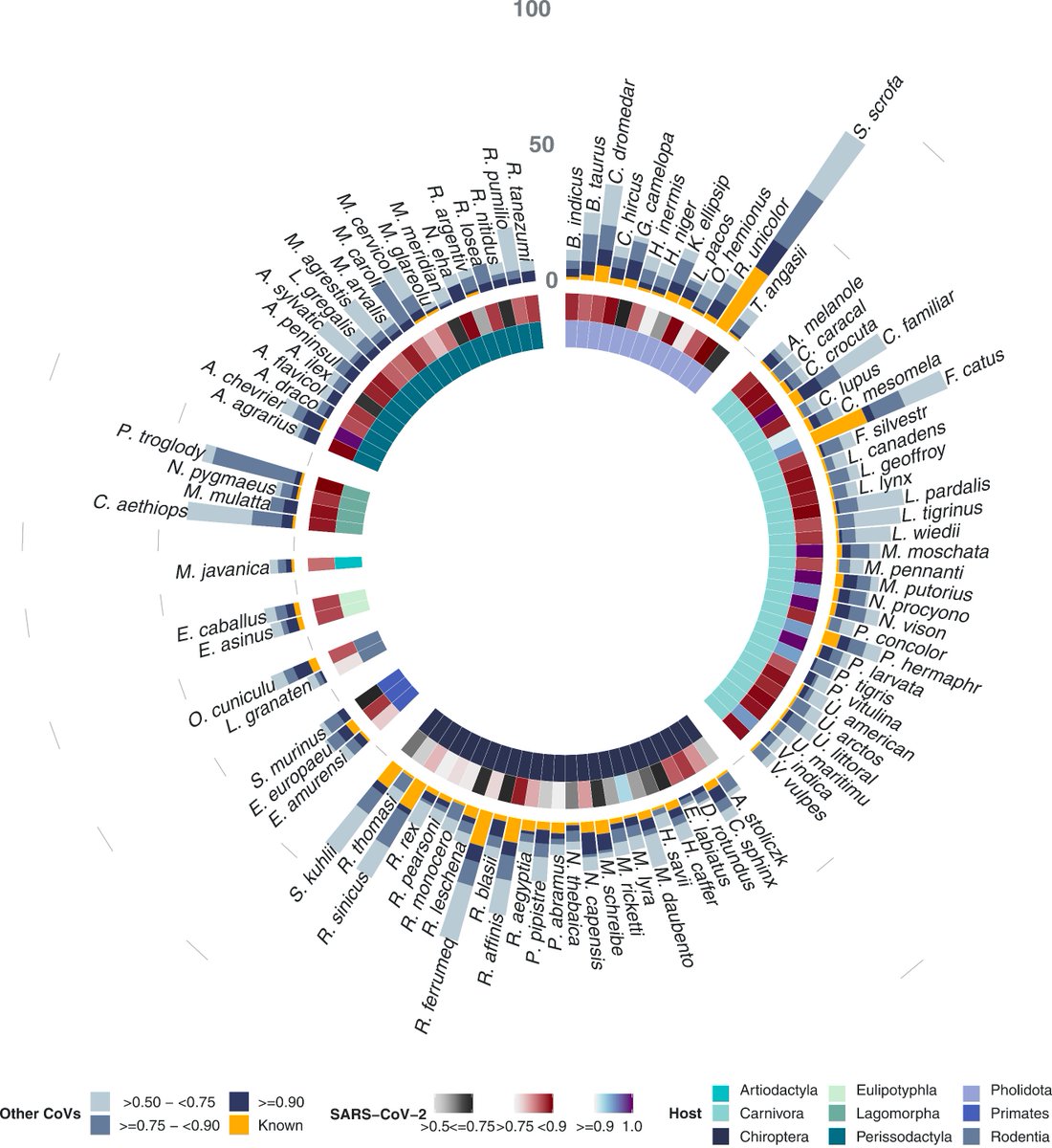
We estimate there are >40 times more mammal species that can be infected with a diverse set of coronavirus strains than was previously known.
We were also able to predict potential recombination hosts of SARS-CoV-2!
10/16
We estimate there are >40 times more mammal species that can be infected with a diverse set of coronavirus strains than was previously known.
We were also able to predict potential recombination hosts of SARS-CoV-2!
10/16
Important:
Viral recombination is distinct from mutations. Recombination occurs over longer periods of time and can generate novel strains or species.
11/16
Viral recombination is distinct from mutations. Recombination occurs over longer periods of time and can generate novel strains or species.
11/16
Limitations:
Our results draw on limited data on coronavirus genomes and virus-host associations. There are also research biases for certain animal species, all of which present uncertainty in the predictions.
12/16
Our results draw on limited data on coronavirus genomes and virus-host associations. There are also research biases for certain animal species, all of which present uncertainty in the predictions.
12/16
Purpose:
Our work can help target surveillance programmes to discover future strains before they spill-over to humans, giving us a head-start in combating them.
13/16
Our work can help target surveillance programmes to discover future strains before they spill-over to humans, giving us a head-start in combating them.
13/16
Future:
We have just been awarded a NERC covid-19 urgency grant to add bird species, therefore, encompassing the full range of important coronavirus hosts.
14/16
We have just been awarded a NERC covid-19 urgency grant to add bird species, therefore, encompassing the full range of important coronavirus hosts.
14/16
Future:
We are also constructing a multiplex species-level contact network, accounting for behaviour and habitat utilisation of host species, to give a broader overview of potential coronavirus associations.
15/16
We are also constructing a multiplex species-level contact network, accounting for behaviour and habitat utilisation of host species, to give a broader overview of potential coronavirus associations.
15/16
Thanks to making it to the end of the thread!
There are more findings, in-depth discussion, and details of methods here:
https://www.nature.com/articles/s41467-021-21034-5
16/16
There are more findings, in-depth discussion, and details of methods here:
https://www.nature.com/articles/s41467-021-21034-5
16/16

 Read on Twitter
Read on Twitter