Tweet thread incoming! Its my first attempt at this so be kind! Our recent paper in @NatureComms shows how we use high resolution AFM and MD simulations together to understand the structure of supercoiled DNA with base-pair resolution... https://www.nature.com/articles/s41467-021-21243-y
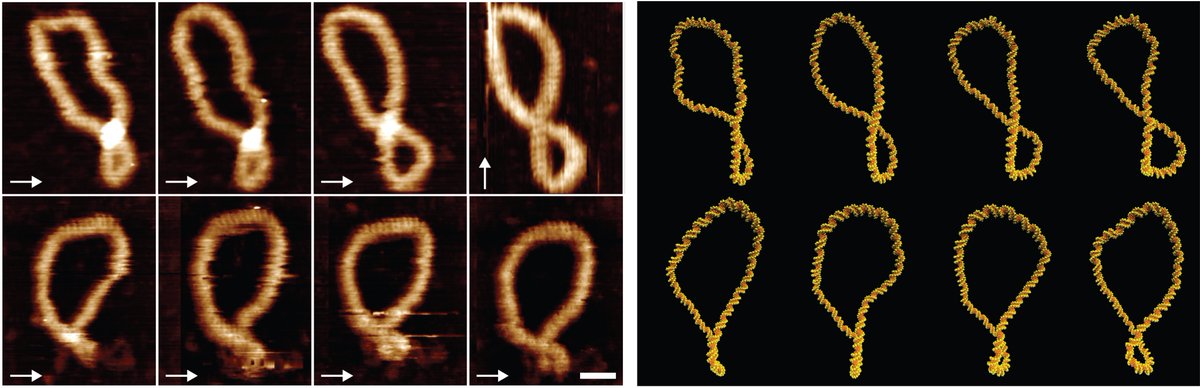
This paper has been a long time in the making! We show that we see complete synergy between AFM images of DNA structure (taken @BrukerNano HQ in SBA), and MD simulations from the incredible @ANoyLab & #notontwitter #SarahHarris
Not only do we see that the structures look the same, but we also see that the dynamics of how supercoiled DNA 'dances' are very similar
We use this similarity to "boost" the resolution of our images to understand how much you can bend DNA before it denatures, just like if you bend a ruler too far, eventually it will snap! We use the simulations to see exactly what's happening with the base pairing at these points
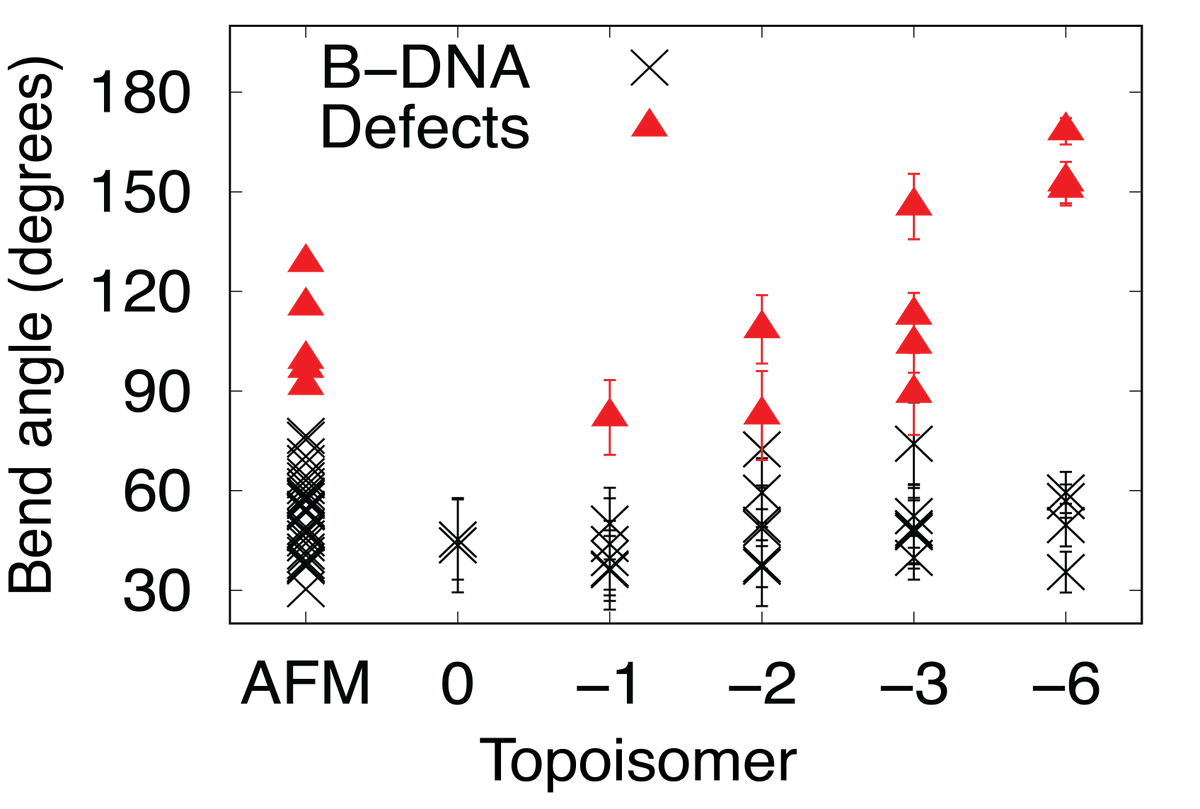
We can correlate these back to the AFM images, using our 'helical' resolution to determine the maximum bending angle for DNA before it denatures
Its here that we see that this denaturation happens more at higher levels of negative superhelical stress. To find out why, we looked at the AFM images...
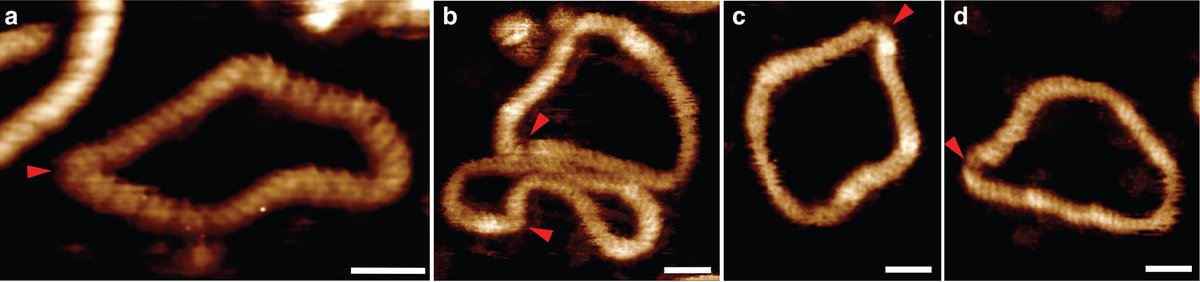
When we looked at individual molecules in the AFM images, we saw that as in the MD the more negative supercoiling we induced, the more defects/bubbles and we saw that this caused much more interesting structures to form
To understand the effect of these defects on the structure of the DNA, we wrote some Python code - TopoStats ( http://www.github.com/AFM-SPM/TopoStats) to automate the analysis of our AFM images and trace the molecules of DNA with help from @JosephBeton and #MayaTopf
This allowed us to find out what's happening to the structure of supercoiled DNA, and this is where it gets really interesting... We see that relaxed DNA is fairly uniform and circular, as expected, but as we increase the levels of supercoiling, the levels of compaction increase
But when we when we get to DeltaLk = -2, the minicircles open up again, as if 'breathing'. It turns out that this the point at which defects form, enabling the DNA to adopt many more shapes, with linear regions stabilised by defects.
(Theres some really exciting synergy with oxDNA results showing plectoneme pinning by defects here https://www.nature.com/articles/srep07655)
This is really exciting because it occurs at a superhelical density close to that found in say bacteria, indicating that DNA may always be on the cusp of being able to form defects, giving it access to a much larger range of 'shapes' for molecular recognition
We demonstrate that this has an effect on recognition by looking at how triplex formation is affected by superhelical density. An aside - it is really quite hard to see triplexes, oligos which intercalate within the DNA double helix, by AFM!
So here @ANoyLab takes over, and uses their incredible simulations to show that actually triplex formation is really able to occur across a wide range of superhelical densities
@ANoyLab shows that triplex formation is both sensitive to supercoiling, but also dealt with by a very clever balance of interactions, to allow triplexes to form across this range of superhelical densities, with almost no change in conformation
This work has been the culmination of years of brilliant collaboration with the DNA stars, many of whom are #notontwitter. Its my first big paper @msesheffield and I couldn't be happier to he here setting up the new @RoyceSheffield Nanocharacterisation Lab with @LunchBobs
This is really special for me, as it sees #notontwitter #KavitMain publish his first paper with me! Thanks for all your hard work and for starting your PhD journey with me supported by @LondonNanotech and @rthoroga who keeps everything going!
Thanks to everyone @BrukerNano for all their support, especially #AndreaSlade #ThomasMueller #BedePittenger #ChanminSu #ShuiqingHu & #DeanDawson over in SBA who helped get these incredible measurements
Thanks to the fab @JohnInnesCentre's @cemstevenson #TonyMaxwell #LesleyMitchenhall #FioreCugliandolo #MichaelPiperakis for the biochemistry and SPR. Special shoutout to @toponano @LivUni for his amazing insight throughout.
And finally, a special thanks to @LynnZechiedrich and #notontwitter #JonathanFogg for pioneering these fantastic DNA minicircles, helping us out, and sending us circles - we couldnt have done it without you! https://www.sheffield.ac.uk/news/first-videos-show-helix-dancing-dna-developed-scientists

 Read on Twitter
Read on Twitter