Our latest preprint examines cross-species prediction of transcription factor binding sites using neural networks. Really wonderful work from Kelly Cochran ( @kellycochra), in collaboration with @divyanshi91, @avshrikumar, @_bakshay, and @anshulkundaje: https://www.biorxiv.org/content/10.1101/2021.02.13.431115v1 1/n
While TF binding sites are not highly conserved between mammals, TF motifs and features of regulatory regions are. It should be possible to train cell-specific models using TF ChIP-seq data from one species, and predict where the TF will bind in the same cell type in another 2/n
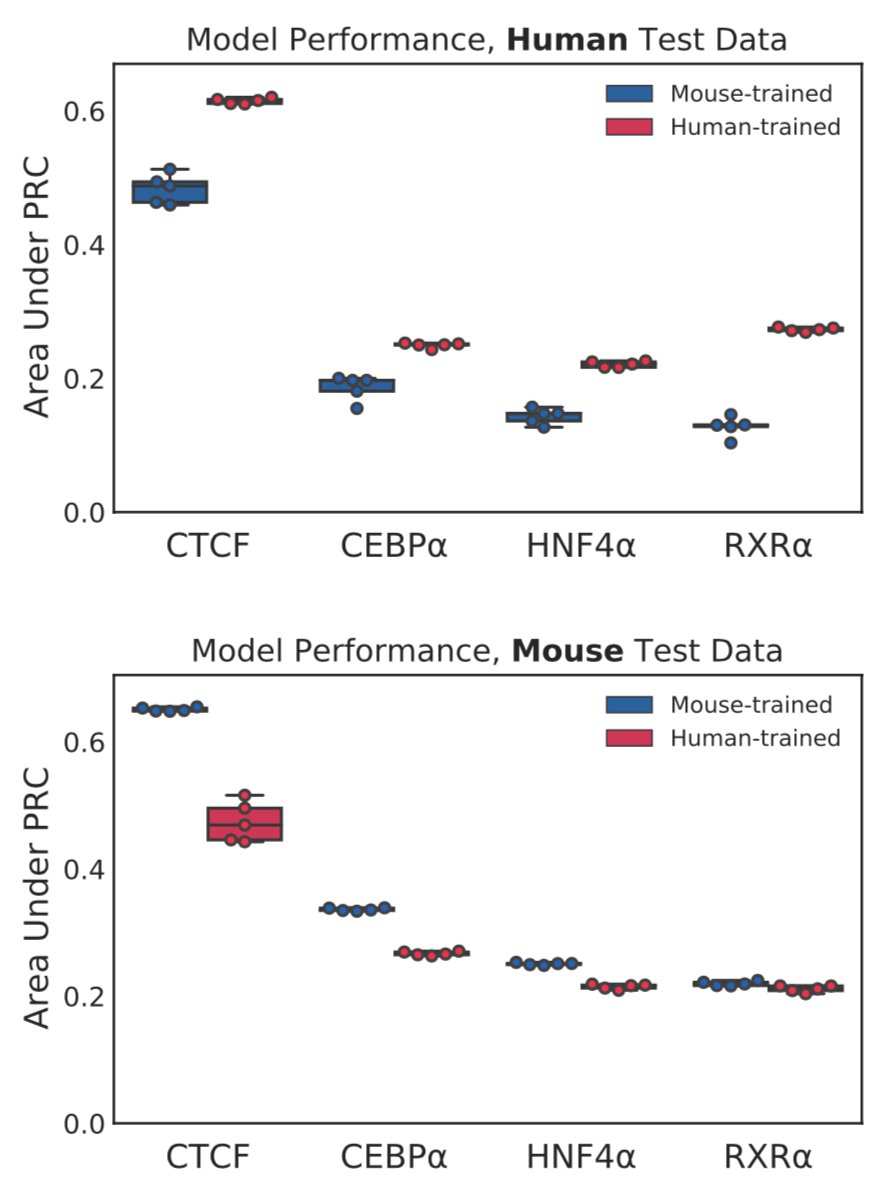
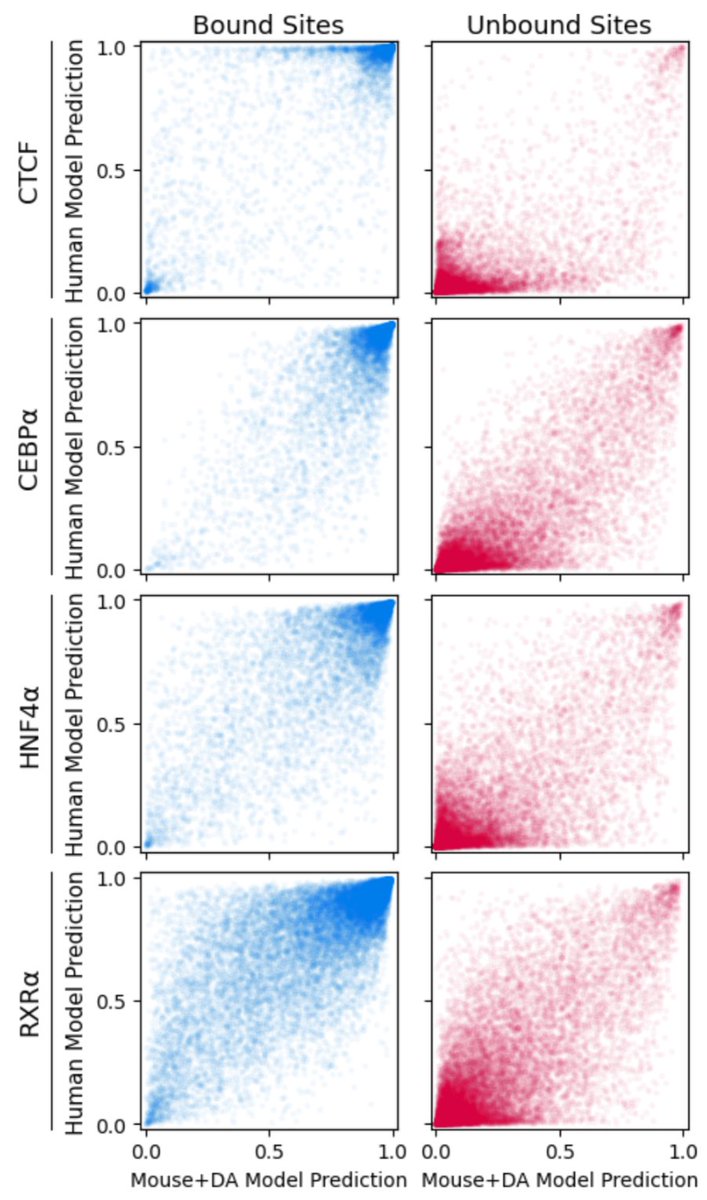
But, as others have in previous work, we find a persistent cross-species performance gap in TF binding prediction. Cross-species NN predictions perform consistently worse than within-species predictions. 3/n
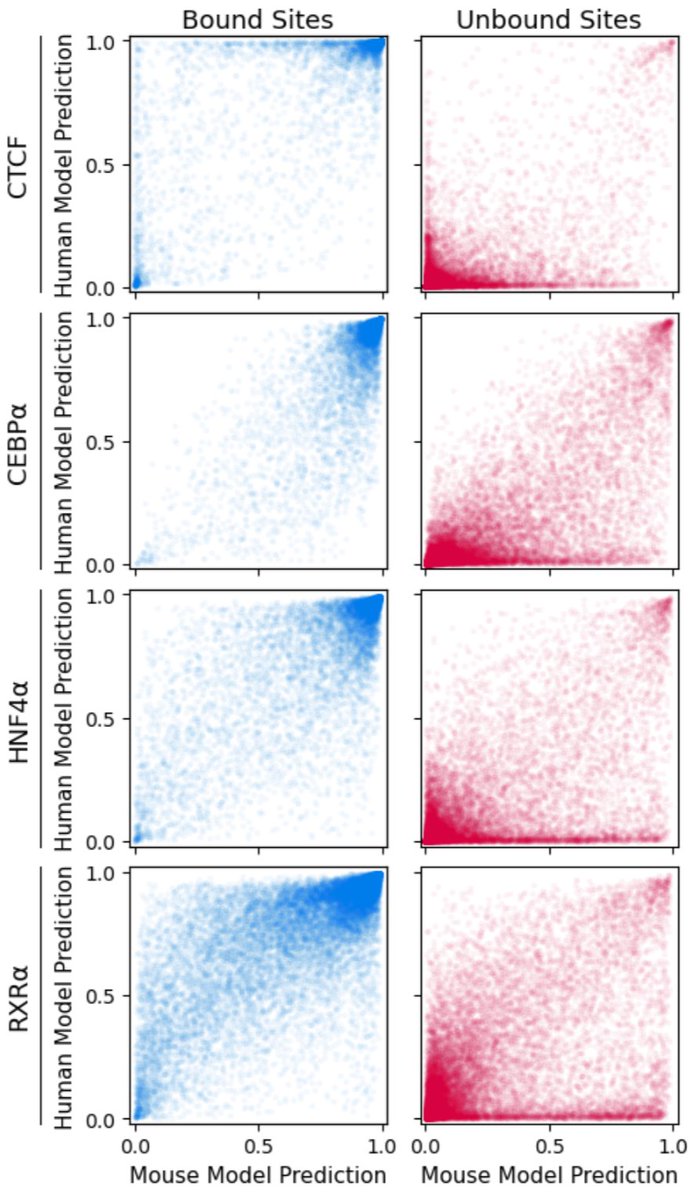
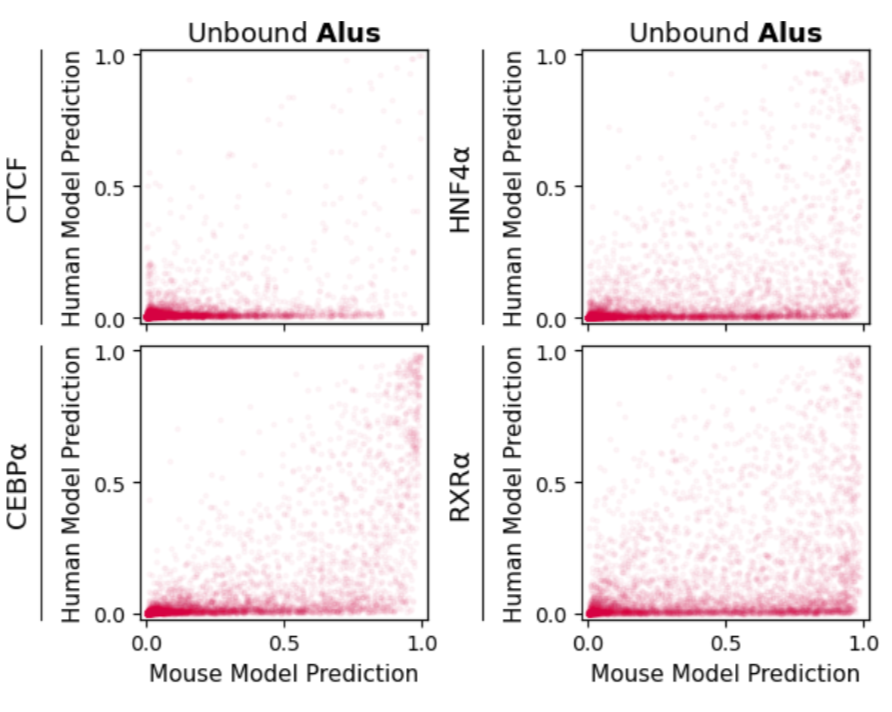
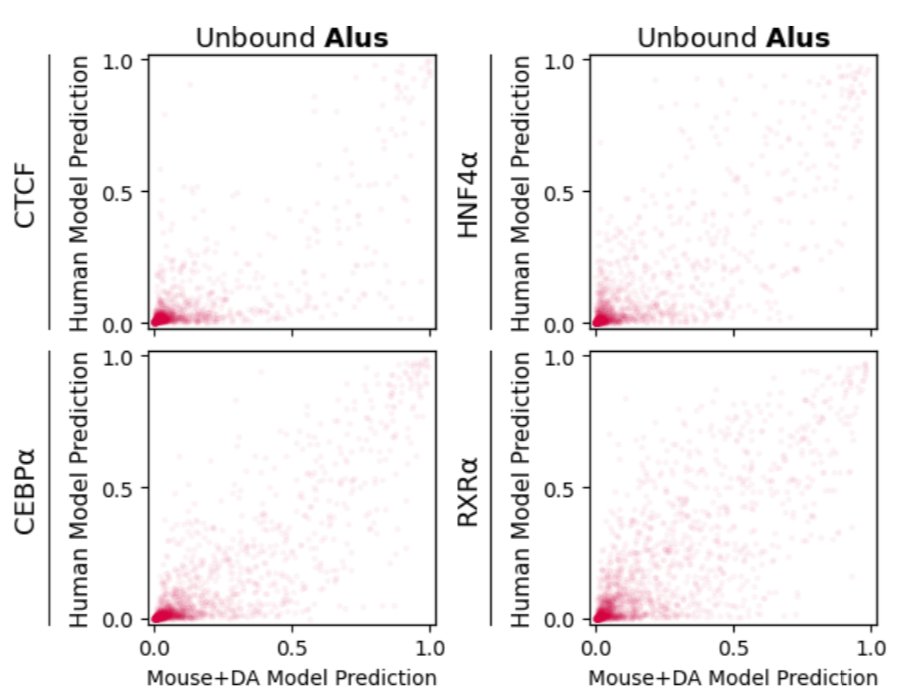
We found that one major source of false-positive predictions is species-specific repeats. Simply put, an NN trained with mouse data has never seen primate-specific repeats, and makes lots of false-positive predictions in the ~1 million Alu elements on the human genome. 4/n
In fact, if you train an NN on human data that excludes SINE elements, you get the same types of false positive predictions in the human genome. 5/n
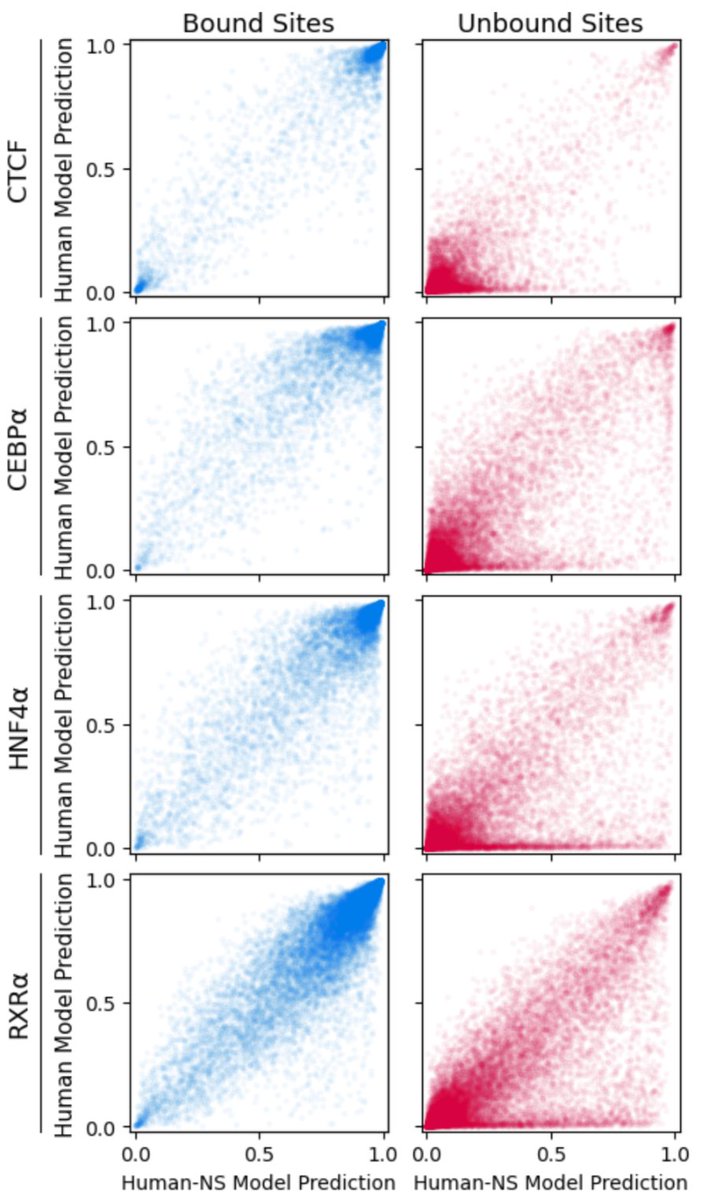
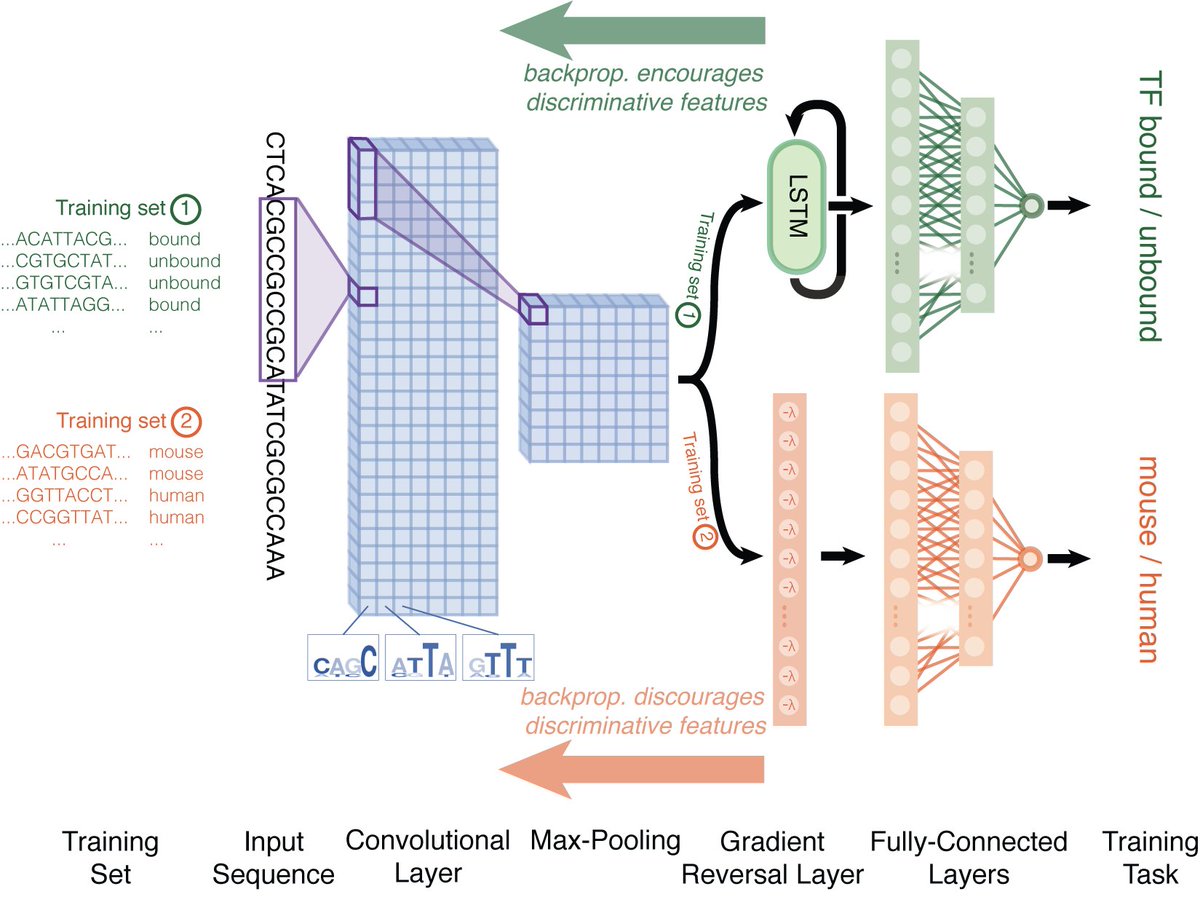
To address this problem, we tried a simple domain adaptation scheme. Half the network trains to predict binding using mouse ChIP-seq data. Half trains on random mouse and human sequences. But a gradient reversal layer *discourages* features that discriminate between species. 6/n
This simple approach solves the misprediction of human-specific repeats! Doesn't solve all cross-species prediction problems, but it's a start. It's straightforward to implement, and doesn't require any knowledge of regulatory regions in the target species. 7/n
We were very fortunate to have the phenomenal @kellycochra work with us during her "gap" year between undergrad and grad school, during which she drove this collab with @anshulkundaje. Kelly is now working with @anshulkundaje at Stanford, and has amazing things in her future! n/n

 Read on Twitter
Read on Twitter