Chance, parallelism or convergence? A inspired by today's preprint. https://www.medrxiv.org/content/10.1101/2021.02.12.21251658v1
inspired by today's preprint. https://www.medrxiv.org/content/10.1101/2021.02.12.21251658v1
 inspired by today's preprint. https://www.medrxiv.org/content/10.1101/2021.02.12.21251658v1
inspired by today's preprint. https://www.medrxiv.org/content/10.1101/2021.02.12.21251658v1
The language and inferences of evolutionary biology have never been more central to our shared dialogue. SARSCoV2 is evolving as the pandemic spreads, & teaching us about this process. Some lineages have gained mutations that increase their fitness (~transmission rate).
In fact the *same* mutation(s) have evolved and spread in multiple independent lineages, in different locations and with their own particular history of prior mutations.
https://www.wired.com/story/worrisome-new-coronavirus-strains-are-emerging-why-now/
How do we describe this phenomenon and what does it mean?
https://www.wired.com/story/worrisome-new-coronavirus-strains-are-emerging-why-now/
How do we describe this phenomenon and what does it mean?
I am an evolutionary biologist who studies how microbes adapt to new environments. Our favorite method is experimental evolution, where we grow multiple independent populations of microbes under identical conditions for days, months, or years. https://msphere.asm.org/content/3/3/e00121-18
We design these conditions to answer questions like how resistance to antibiotics evolves, how new hosts are colonized, how diversity itself arises. e.g. https://pubmed.ncbi.nlm.nih.gov/32457248/ @shellyscrib @SirMicrobe @ASantosLopez
We use multiple replicate populations to essentially replay the tape of life because evolution is subject to random forces like mutation and genetic drift, as well as deterministic forces like natural selection. This study in mice made this clear! https://pubmed.ncbi.nlm.nih.gov/32398278/
One result that we eagerly anticipate is the *same new trait arising in multiple independent populations.* We often don’t know what this trait will be at the outset, but the complete genome sequences of the evolving populations can point the way.
If the same gene or even the same amino acid repeatedly changes, independently, we have a strong inference that that change is an adaptation. How do we describe this phenomenon? Is it “parallel evolution?” Is it “convergent evolution?” Does it matter? https://www.annualreviews.org/doi/10.1146/annurev-ecolsys-110617-062240
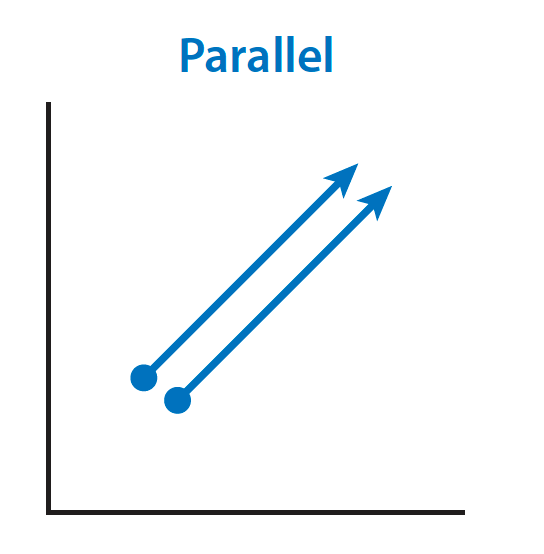
Some definitions. Parallel evolution is the evolution of similar phenotypes or genotypes in multiple independent populations, in response to similar selection pressures, from *similar initial conditions.* Here’s a schematic:
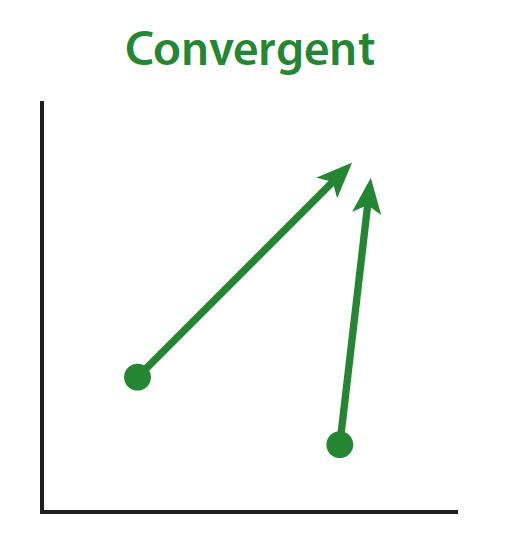
Convergent evolution is the evolution of similar phenotypes or genotypes in multiple independent populations, in response to similar selection pressures, from *different initial conditions.* Here’s another schematic:
The key distinction is how we quantify similarity of starting conditions. Are starting pop's identical but growing in different envts or hosts? Are they genetically different but growing in same condition? For SARS-CoV-2, which just 15 mo ago was 1 virus in 1 host, it's tricky!
Our new article used “parallel” and “convergent” somewhat interchangeably . In the US, we found at least 7 genetically independent lineages w/ a new mutation at Spike amino acid 677, which is Q in the original SARS-CoV-2 as well as related bat CoV. https://www.medrxiv.org/content/10.1101/2021.02.12.21251658v1
. In the US, we found at least 7 genetically independent lineages w/ a new mutation at Spike amino acid 677, which is Q in the original SARS-CoV-2 as well as related bat CoV. https://www.medrxiv.org/content/10.1101/2021.02.12.21251658v1
 . In the US, we found at least 7 genetically independent lineages w/ a new mutation at Spike amino acid 677, which is Q in the original SARS-CoV-2 as well as related bat CoV. https://www.medrxiv.org/content/10.1101/2021.02.12.21251658v1
. In the US, we found at least 7 genetically independent lineages w/ a new mutation at Spike amino acid 677, which is Q in the original SARS-CoV-2 as well as related bat CoV. https://www.medrxiv.org/content/10.1101/2021.02.12.21251658v1
In 6 lineages this Q mutated to H, but remarkably in 2 different ways at the nucleotide level. In the 7th lineage, Q-> P. The parents of these lineages differ by a handful of mutations each, but they share some attributes also, including the well-described D614G mutation in Spike
From the perspective of this 614G mutation, we might call these changes *parallel,* but from the perspective of other mutations, we might call them *convergent.* I don’t think the language matters here but invite discussion.
Perhaps more striking, these aren’t the only 677 lineages of potential concern. 677H has arisen multiple times elsewhere in the world, including Egypt, Denmark, and India. Further, a
newly designated lineage, B.1.525, carries S: Q677H plus
several mutations seen in B.1.1.7.
newly designated lineage, B.1.525, carries S: Q677H plus
several mutations seen in B.1.1.7.
Also, a 19B cluster with the ostensibly less fit, ‘ancestral’ D614 Spike, which has been circulating at ≤ 2% of global frequency since August 2020, recently resurfaced as a newly reemergent lineage carrying N501Y together with 677H. @firefoxx66 @nextstrain
What’s the TLDR? Finding multiple lineages with increasing freq w/ the same mutation is likely not just recurrent chance mutation, but rather a product of chance AND selection. Whether you call this evolution parallel or convergent is up to you. (one more)
Regardless, let’s keep an eye on it and figure out how it works. It’s still a grail of evolutionary biology and evolutionary medicine not just to find new adaptations but also to figure out how they work. Thanks for reading!

 Read on Twitter
Read on Twitter