What does it take to generate multiple cell states? To find out, we designed and built "MultiFate", a synthetic circuit that generates multistability in mammalian cells. https://www.biorxiv.org/content/10.1101/2021.02.10.430659v1 . Work from brilliant student @RonZhu2015 w/ @jesusdelrio_7 and @jgojalvo.
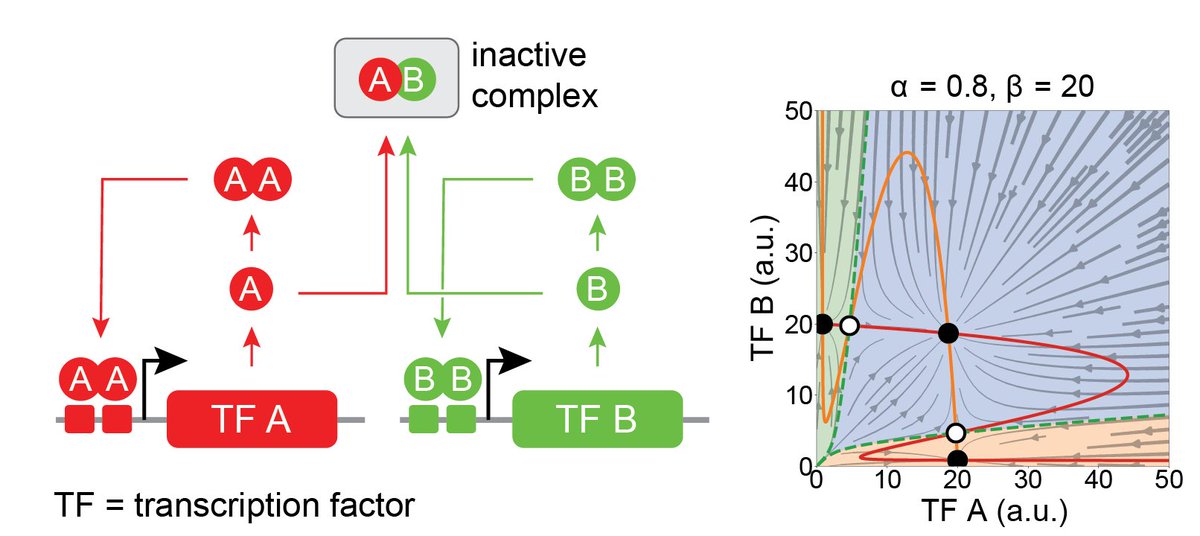
MultiFate is a naturally-inspired design in which transcription factors homodimerize to positively autoregulate their own expression, and heterodimerize to cross-inhibit each other. Here’s a simple example, in which 2 transcription factors generate tristability.
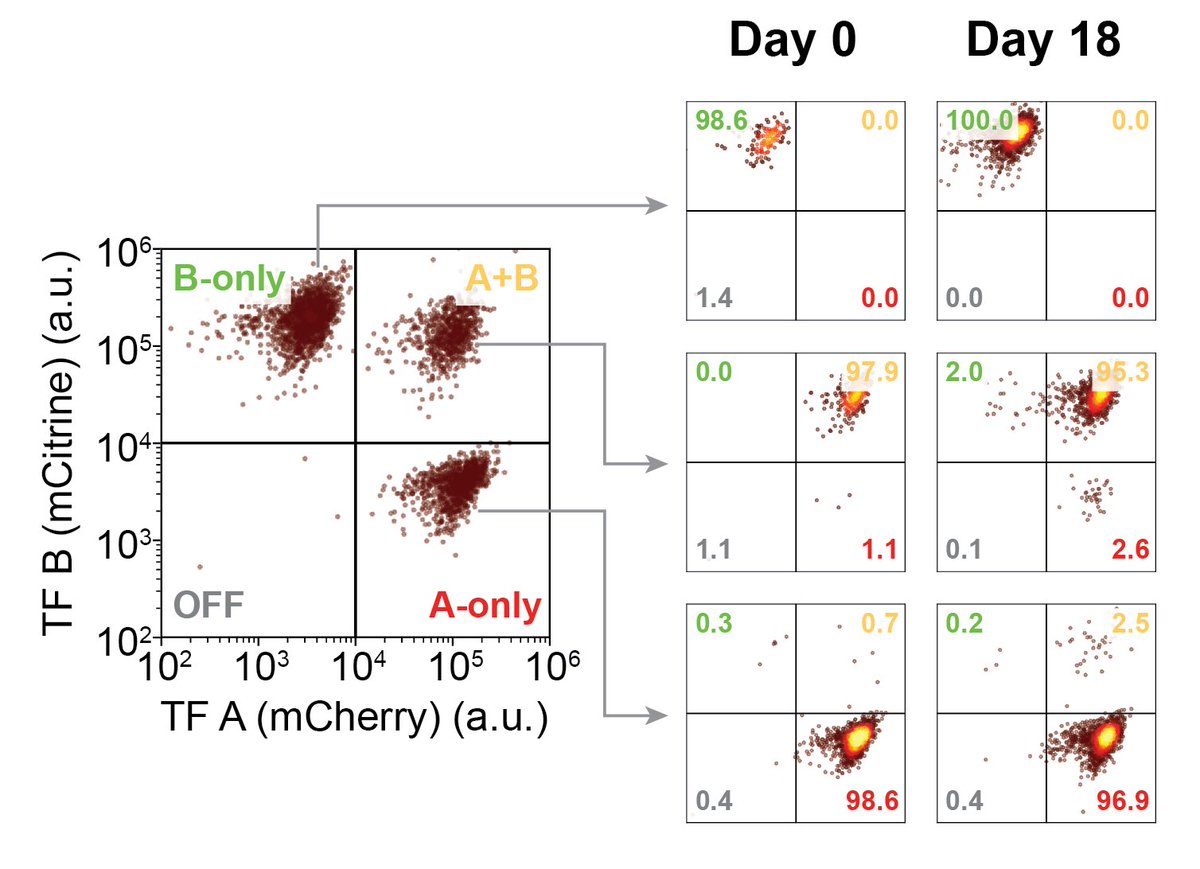
To make MultiFate, we engineered a set of dimer-dependent TFs. The minimal MultiFate system with two of these TFs successfully generates three states, each stable for >18 days, nicely matching model prediction.
MultiFate allows de- and re-stabilization of states. Preinstalled control knobs in TFs allow one to specifically destabilize the ‘double positive’ state by reducing TF half-life, causing bifurcation and irreversible cell transitions, analogous to natural differentiation.
MultiFate scales. Plugging a 3rd transcription factor into the 2 TF system generates 7 states (septastability), with no re-engineering of the existing circuit (because inhibition occurs at the protein level.) All 7 stable for >18d. # of states grows exp w/num of TFs.
Stability of these 7 states can be vividly visualized in time-lapse movies. Here is movie of cells with the 3 TF circuit. Individual cells in each of 7 states grow into colonies retaining initial state. TFs are shown in RGB, so each of the 7 states is a distinct color.
MultiFate can be the basis for synthetic mammalian circuits that use multiple cell states, just like natural systems do.
This work builds on elegant work engineering zinc finger TFs by @Jlohmueller, @MoKhalilLab, @timlu, Caleb Bashor, James Collins, Pamela Silver & others.
This work builds on elegant work engineering zinc finger TFs by @Jlohmueller, @MoKhalilLab, @timlu, Caleb Bashor, James Collins, Pamela Silver & others.

 Read on Twitter
Read on Twitter