1/ [Long! Tweetorial] I'm excited to finally share the result of the largest project of my PhD. Together w the paper we published a year ago, this paper helped serve as a proof-of-concept for starting @Dyno_Tx. https://www.nature.com/articles/s41587-020-00793-4#author-information
2/In 2015, when @ekelsic joined the @geochurch lab, he was looking for opportunities to marry emergent high-throughput synthesis technologies with #machine_learning.
3/ The key insight was to focus on a protein that is hard to model biophysically, but if modeled has a large impact and not just academically interesting. Basic science, engineering, and useful outcome together.
4/AAVs check the boxes, because they are very promising gene therapy vectors. The idea was to learn to predict their functionality directly from data. I joined this group sometime in 2016, and shortly after we had an opportunity to start a collaboration with @GoogleAI.
5/In particular shoutout to Drew Bryant, Ali Bashir and @lucycolwell37 our patient collaborators for many years. We worked on this project together for 3 years, meeting remotely every week before it was cool, to study how to best use ML for various properties of interest in AAV.
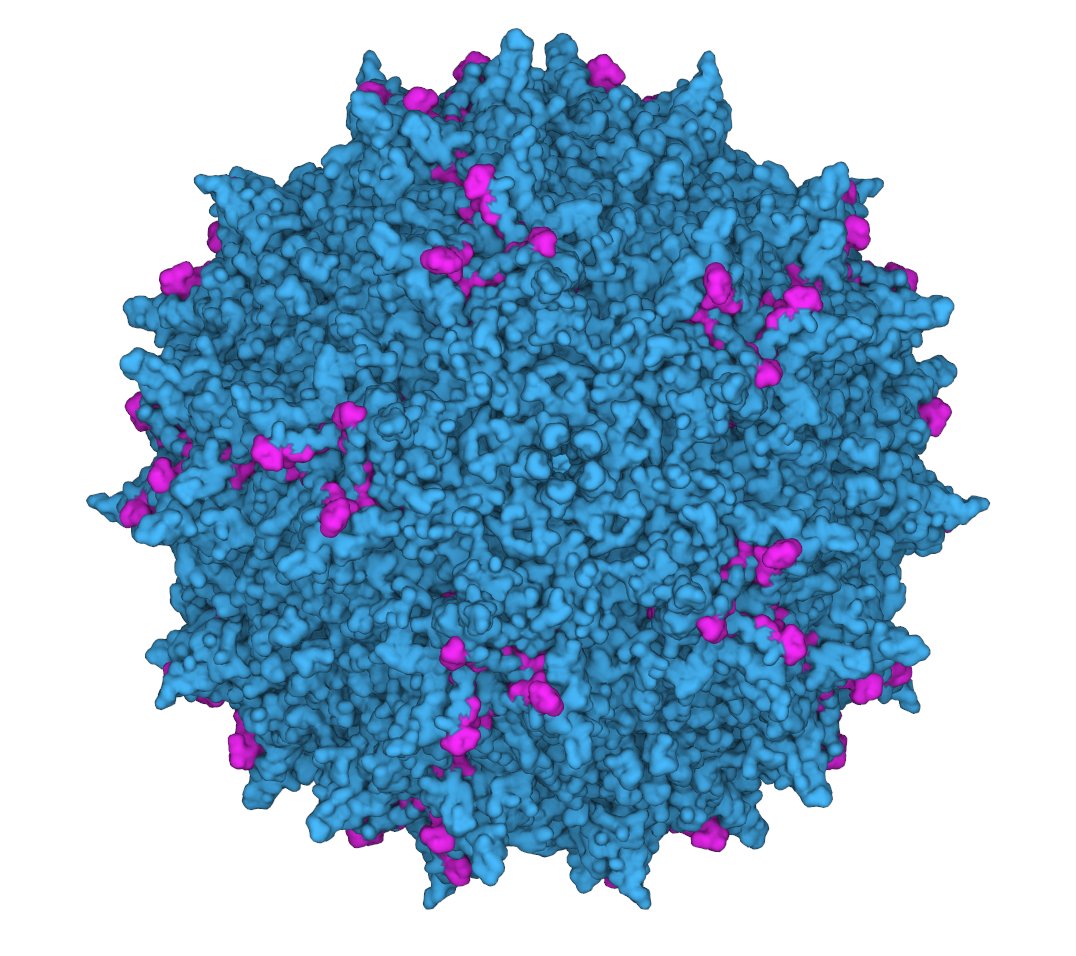
6/ We targeted a 28 amino-acid region of the AAV2 capsid protein, which was relevant for multiple properties and had both buried and surface-accessible residues. This size was partially imposed by limitations in DNA synthesis length at the time.
7/An important problem (less obvious in hindsight), was that at the time we had no idea if ML models will be able to predict anything beyond a few mutations away. Most of the work in literature was focused on validating ML algorithms on #mutational_scans with low edit distances.
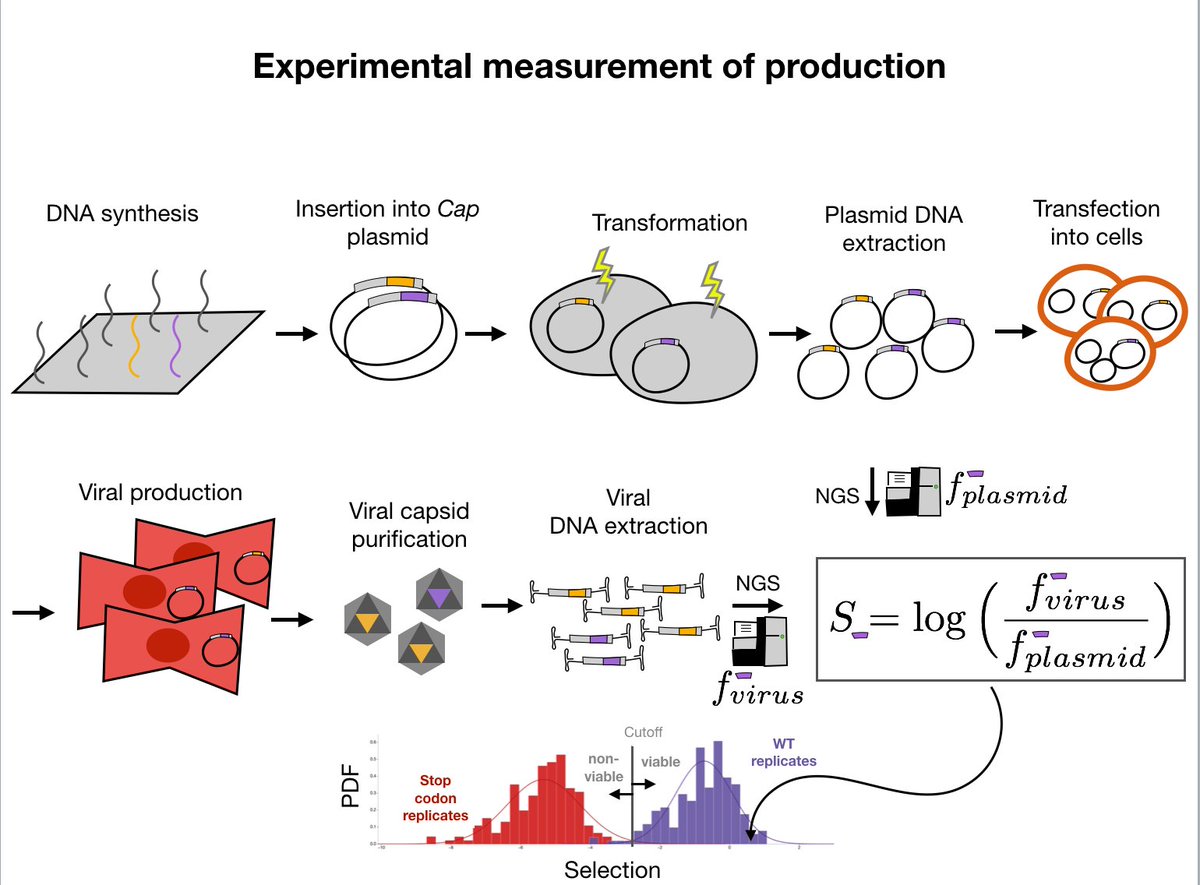
8/ We decided to focus on the property with the cleanest signal, packaging efficiency, which in binarized form we call "viability". If the virus packages as well as the WT or better we call it viable, otherwise we call it "non-viable".
9/With this property as our training objective, we used a dataset of 70000 sequences that we had collected using random samples and a linear model (details in paper) to train logistic regression, CNN, and RNN models.
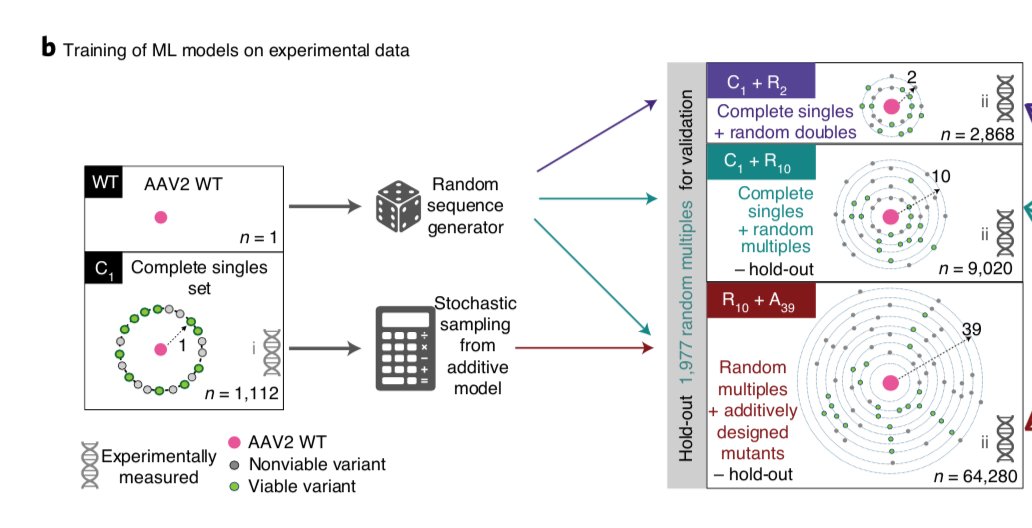
10/But the training dataset was generated in a biased manner, we also decided to study the role of data distribution on the performance of the models. We used three partitions, single and double mutants only (Potts-model inspired), randomly generated mutants, and the full data.
11/Our models seemed to work decently on holdout, but we had very few randomly sampled viables beyond edit distance 5. While we did find numerous viable mutants up to distance 21 (counting insertions) in our full dataset, that wasn't in our holdout due to its generation bias.
12/ One problem was to use these one directional models to extrapolate, i.e. predict viable variants beyond the edit distance in our dataset. We designed sequences in two ways.
13/ First, we just randomly sampled 2.5 billion sequences in-silico in the space around the wildtype and scored them with each model, keeping each model's favorites.
14/Second, we decided to let the model manipulate the initial seeds to improve it with a simple (deterministic) greedy algorithm and tried going deep into the sequence space. It felt risky then (what if nothing validated?), but in retrospect we could have pushed it even further.
15/We made 200K sequences, and measured their packaging ability. What did we learn? First, it is quite hard to find anything good more than 15 mutations away by random sampling, even if you vet them with a model. It's much better to use the model to guide you to the right place.
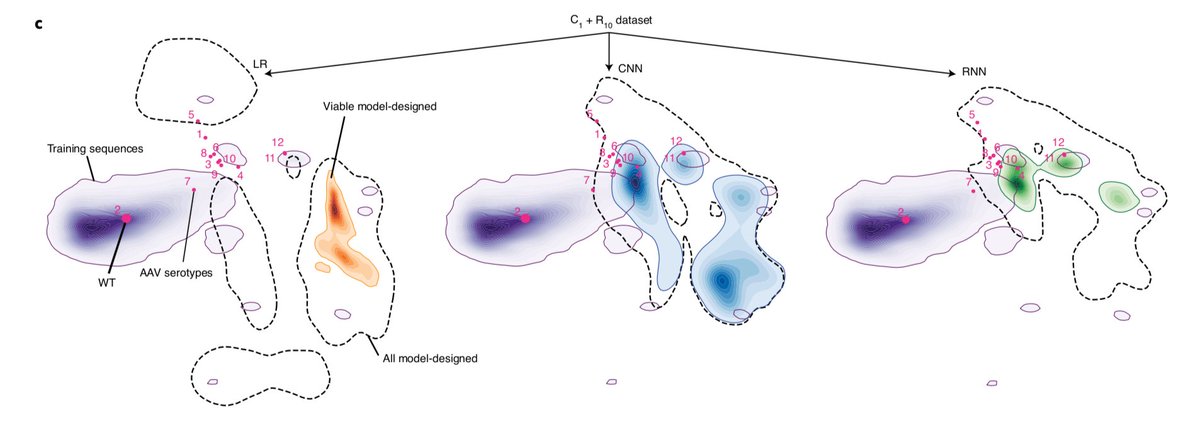
16/LR model did particularly well with one of the training sets. This was somewhat surprising. But in retrospect it made sense, it found a single peak and walked everything there. As a model it was highly sensitive to input data distribution.
17/ LR also rescued most inviable seeds (Fig S5). The neural networks tended to be more robust to the input data. CNNs were more focused on positional changes, while RNNs tended to switch between AAs with similar properties. Both worked better than LR with non-random data.
18/Overall, we found that apart from diversity in the "deep" sense (how far into the sequence space we can go), CNNs tended to span a larger volume of viable capsids. The best LR was quite narrow in this sense (see figure 4 for details).
19/There is a lot that can be done with this data, some we have explored, but much we have left for future work by us or others. We look forward to seeing how people use this data to gain insights about ML for protein design.
20/Tying it back to #AAV s, one aim that we have is to diversify AAVs enough that we can circumvent pre-existing immunity and improve tissue targeting for #GeneTherapy.
21/ Since this study was conducted, DNA synthesis has come a long way, and we also have done a lot of work @Dyno_Tx to build better models and design algorithms. Using these, we are designing AAVs with far more complex phenotypic profiles, and testing them in-vivo.
22/If this work gets you excited, you should reach out to us to talk science, or better, join us at Dyno for this adventure! We are growing fast, and there are so many cool problems to solve that are scientifically interesting, challenging, and impactful. https://www.dynotx.com/careers/

 Read on Twitter
Read on Twitter