I am delighted to share with you our work from the @MundlosLab at @MPI_MolGen that just came out in @Nature.
We report that deletions involving a previously unannotated long non-coding transcript cause the loss of ventral limb identity during development by regulating En1 expression specifically in the limb. This results in so far undescribed severe congenital limb malformations.
Below, you find a full thread and for more details check out our work here: https://www.nature.com/articles/s41586-021-03208-9
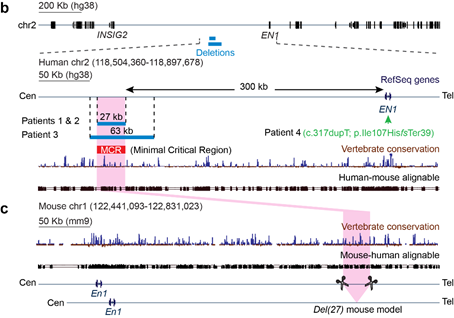
Clinical researchers from Brazil and India first characterized the disease in patients with severe congenital limb malformations including syndactyly, ventral nails, triangular fibulae, and triangular tibiae. Strikingly, the knees of the patients are not oriented to the front!
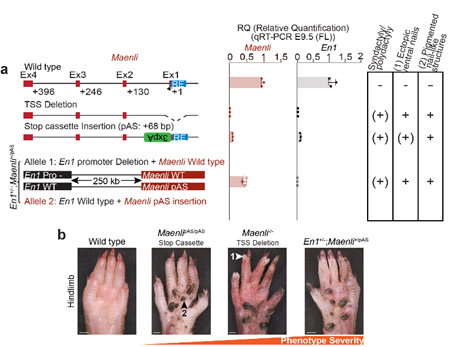
Their DNA samples were sent to Andrea Superti-Furga from the University of Lausanne in Switzerland for genetic testing. There, the team of Human Geneticists identified homozygous deletions of about 27 kb in a large gene desert of about 700 kb.
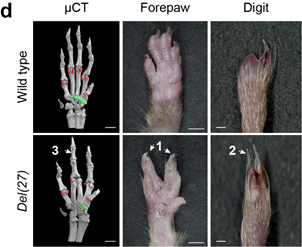
At @MPI_MolGen, we produced an animal model for the newly identified disease by deleting the homologous region in mouse. The mouse model partially recapitulated the human phenotype with the presence of the characteristic syndactyly and ventral nails.
Interestingly, this phenotype was similar to the previously reported engrailed-1 knockout phenotype. However, while En1-/- mice exhibit limb, brain, sternum and ribs defects, our mutants exhibit only a limb phenotype (similar to the patients with an isolated limb phenotype).
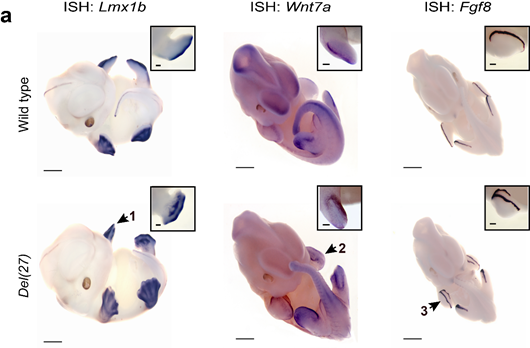
This was consistent with the whole mount in-situ hybridization results with a complete loss of En1 expression specifically in the limb. En1 expression was maintained in the somites and in the mid-hindbrain boundary.
Moreover, the dorsalizing genes Lmx1b and Wnt7a were ectopically expressed in the ventral side of the limbs and the AER marker Fgf8 exhibited a proximo-ventral expansion resembling a double AER. The mice exhibited a double dorsal limb phenotype (loss of ventral limb identity).
Next, we investigated the mechanism by which the deletions caused a complete loss of En1 expression in the limb. First, we hypothesized the presence of DNA regulatory elements within the deletion.
We used available epigenetic data and identified one putative enhancer element within the deletion. We tested its enhancer activity using a transgenic mouse at E11.5 but did not see any detectable enhancer activity in the limb at this stage (data not shown in the manuscript)!
We then hypothesized the presence of a cluster of limb enhancers with redundant activity. However, at the same time, something else retained our attention at the locus. It was the presence of an EST…
This information is often ignored in the interpretation of genetic variants but in this case, it was the key to solving the disease mechanism. #Genetics
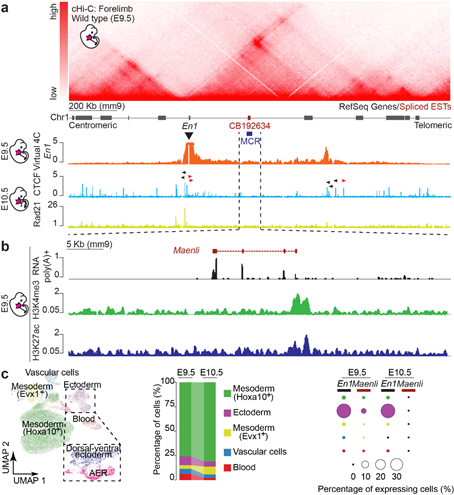
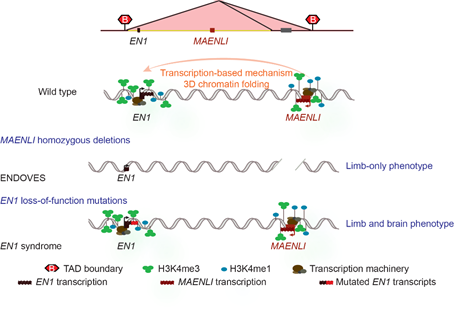
Indeed, based on this, we hypothesized the presence of a putative non-coding transcript at the locus and confirmed this using poly(A)+ RNA-seq and RACE PCR. We named this 4-exon non-coding transcript Maenli (for Master activator of engrailed-1 in the limb).
Next, we examined whether the Maenli locus was required for En1 expression regulation and thus causal for the limb malformations. To test this, we inactivated Maenli transcriptional activity by inserting an early stop cassette (SV40 poly(A)) at the locus.
This significantly altered En1 expression in the developing limb bud and the mice exhibited the characteristic double dorsal limb phenotype, suggesting that the Maenli locus is indeed causal for the limb malformations.
We also investigated Maenli’s mechanism of action and showed that it acts in Cis and not in Trans, and that having a full intact Maenli sequence is not required for its function.
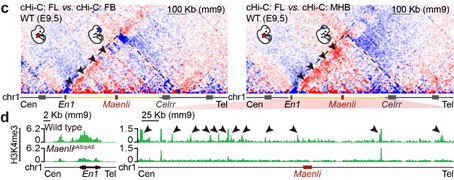
Finally, we investigated its effect on 3D chromatin folding and the epigenetic signature of the En1 TAD and showed that Maenli transcriptional activity helps to establish a permissive chromatin environment at the En1 locus and its surrounding regulatory landscape.
Last, but not least. Following these findings, the clinicians identified an additional patient with a limb phenotype similar to that observed in the above-mentioned patients but, in addition, this patient exhibited severe congenital malformations of the brain.
Interestingly, sequencing of EN1 exons in this patient revealed the presence of a homozygous frameshift mutation recessively inherited from the parents.
Thus, within the same story and for the first time after its discovery in the mid-90s, we involve the EN1 gene and its limb-specific regulatory information (MAENLI locus) in severe congenital malformations with distinct phenotypes.
We also identify a key epigenetic mechanism that regulates En1 expression specifically in the limb and show that it is required for ventral limb specification during development.
This work would not have been possible without our collaborators! I am thankful to all the clinicians from Brazil and India that identified the patients and the Human geneticist from Lausanne especially Andrea Superti-Furga, Carlo Rivolta, Sara Balzano and Mathieu Quinodoz.
I am also thankful to our fantastic colleague Andreas Magg @MioMaggi, a talented PhD student in our lab who contributed to this work. I would also like to thank @StefanMundlos and Phillip Grote @GroteLab1 for their ultimate help and support but also all the authors of this work!
This was a very nice scientific adventure with the amazing @MundlosLab! I will close this thread by thanking the patients and their families, without your consent, this work would not have been possible… simply thank you!
#RareDiseaseMonth #SupportRareDiseaseResearch
#RareDiseaseMonth #SupportRareDiseaseResearch

 Read on Twitter
Read on Twitter