Very excited to see my work with @Simmonds_Lab, Oliver Pybus @EvolveDotZoo, Aris Katzourakis @ArisKatzourakis finally out on bioRxiv -- long overdue mainly because 2020 happened!
I will try to explain the principal behind how the model works.
A thread!
1/ https://twitter.com/PhyDyn_Papers/status/1359421148165857280
I will try to explain the principal behind how the model works.
A thread!

1/ https://twitter.com/PhyDyn_Papers/status/1359421148165857280
The main challenge to molecular dating of viruses is that if we extrapolate their long-term evolutionary trajectory based on their short-term substitution rates, the viral genome sequence should be completely unrecognisable over thousands of years...but they aren't! 2/
In this paper, we tried to develop a mechanistic framework to address this dilemma; inspired by the idea that the primary driver of sequence change in viruses over longer time scales is host adaptation ( https://www.nature.com/articles/s41579-018-0120-2) 3/
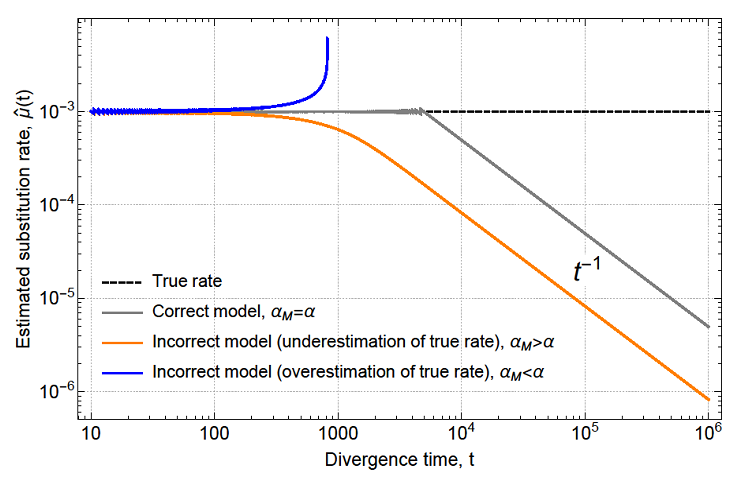
The principle behind the model is simple: chronological saturation of sites (superimposed substitutions), from the fastest evolving to those that evolve epistatically, to those evolving at the host substitution rate, determines the time-dependent evolutionary rate in viruses. 4/
So the basic idea behind saturation of sites is: assuming a constant and uniform substitution rate across all sites, once a pair of sequences have diverged from each for a long time (t*~1/substitution rate), you'd lose all the information to reconstruct their phylogeny. 5/
Because you're essentially reaching the maximum level of differences between the pair, any attempt to estimate substitution rate would be strongly correlated with the timespan of observation (i.e. a power-law rate decay with slope -1 will emerge) 6/
I must say, most of the substitution models are 'smart enough' to distill such effects (to a certain degree!). There are also statistical tests for saturation. I'd highly recommend reading this great book by @DrSimonHo that just came out to learn more about dating species. 7/
BUT, what if there's a large observation gap between the ancestral and derived sequences (i.e. a very long internal node in the tree) and that during this period a substantial fraction of sites have already started saturating? 8/
We think that this is what drives the time-dependent rate effects over long time scales. Our model recapitulates the empirical observation of a universal power-law rate decay with slope -2/3 that Aiewsakun and Katzourakis ( https://jvi.asm.org/content/90/16/7184) found across virus species. 9/
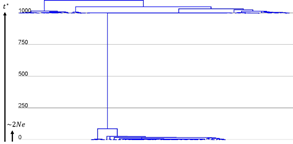
The interesting thing about our model is that we can now re-date the common ancestor of virtually all virus species mechanistically and provide a radical revision of their evolutionary timescales. e.g. here's what the PoW-transformed tree for HCV and Sarbecovirus looks like! 10/
OK, this thread is already getting too long! I think I will stop here and am very keen to hear your thoughts/comments about our work. Thanks for reading all the way down to this last tweet! End/

 Read on Twitter
Read on Twitter