We presented preliminary results of a continuous vector field approach, dynamo, a while ago. Now I am proud to tell you that we have established the full-fledged theory, and proved its broad utility, including app to COVID-19. See a full new version here: https://www.biorxiv.org/content/10.1101/696724v2
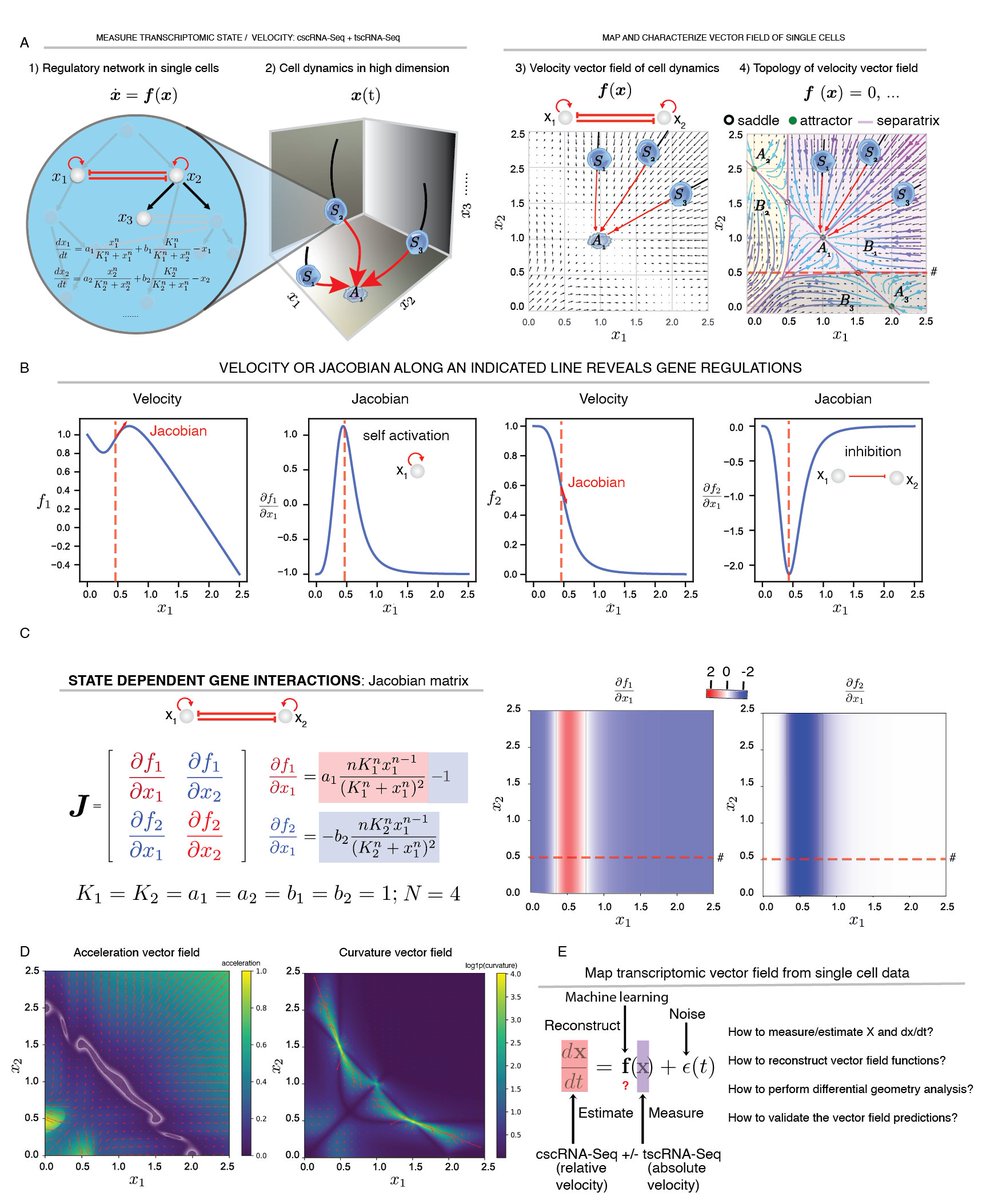
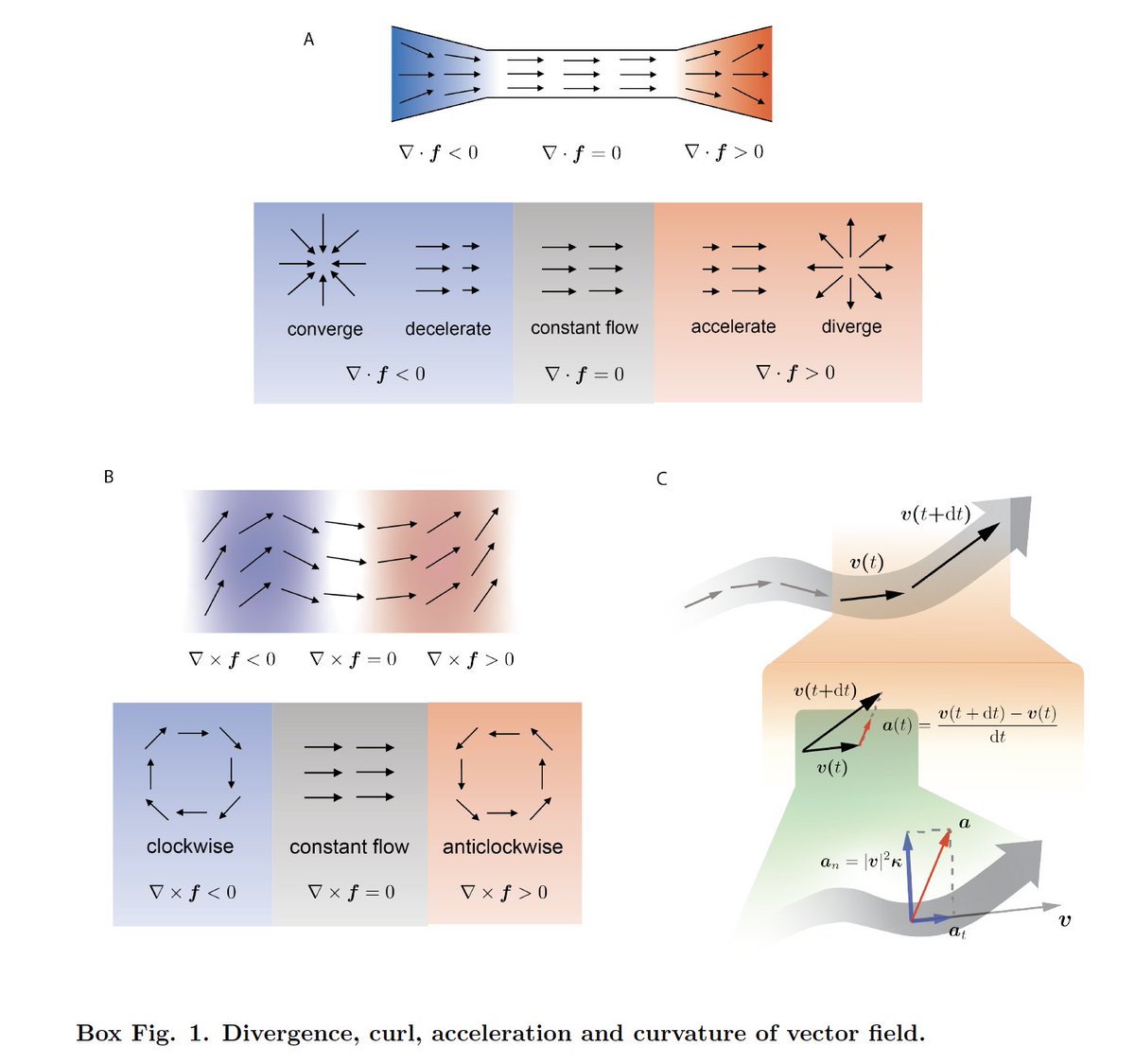
We first introduce key concepts in dynamical systems and differential geometry to single cell genomics: e.g. topology of vector fields predicts stable/bifurcating phenotype while Jacobian state-dependent regulation. We also extend RNA velocity to acceleration/curvature fields
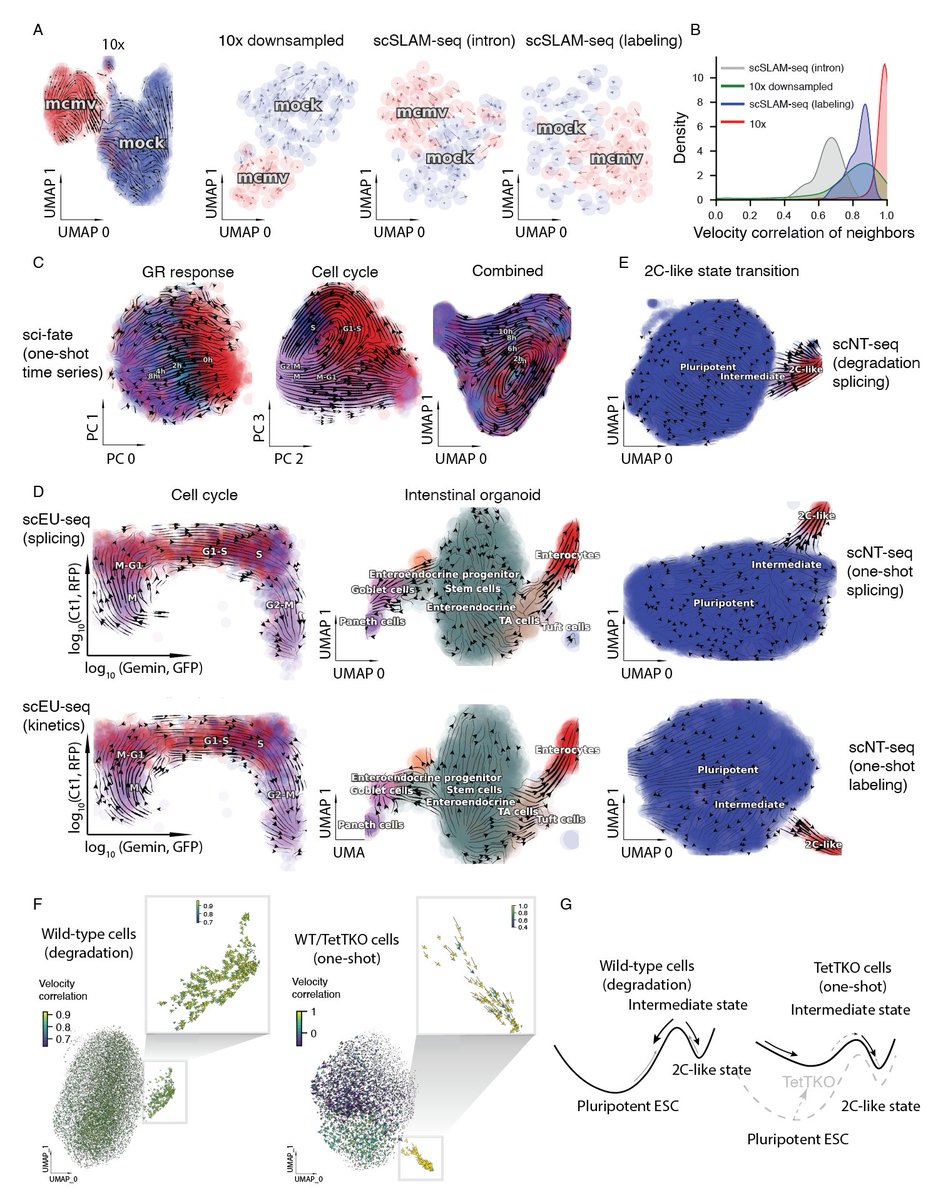
We build an inclusive model of expression dynamics that reconciles splicing and labeling kinetics. This framework generally applies to all metabolic labeling based scRNA-seq (tscRNA-seq) to obtain absolute turnover rates, revealing faster degradation in the mouse than human.
This model also gives rise to absolute instead of relative spliced RNA velocity, and unspliced, new and total RNA velocity, helping us to deconvolve glucocorticoid responses from orthogonal cell-cycle progression, revealing mechanism of pluripotent to 2c-like state transitions.
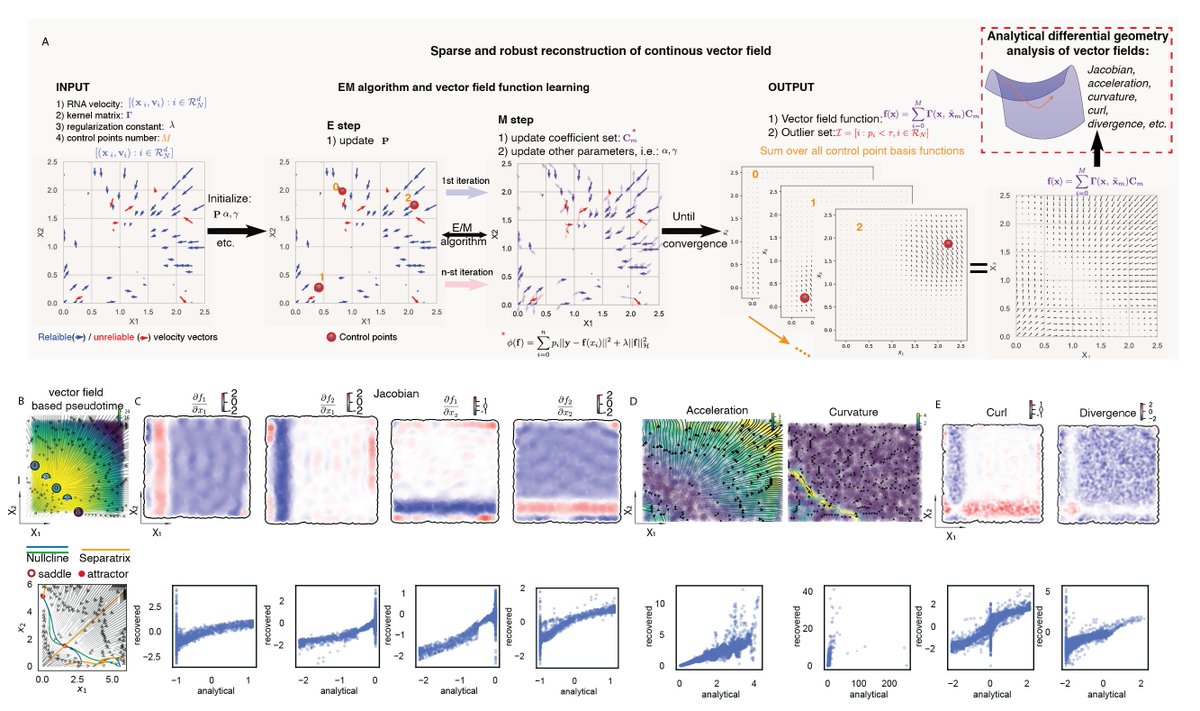
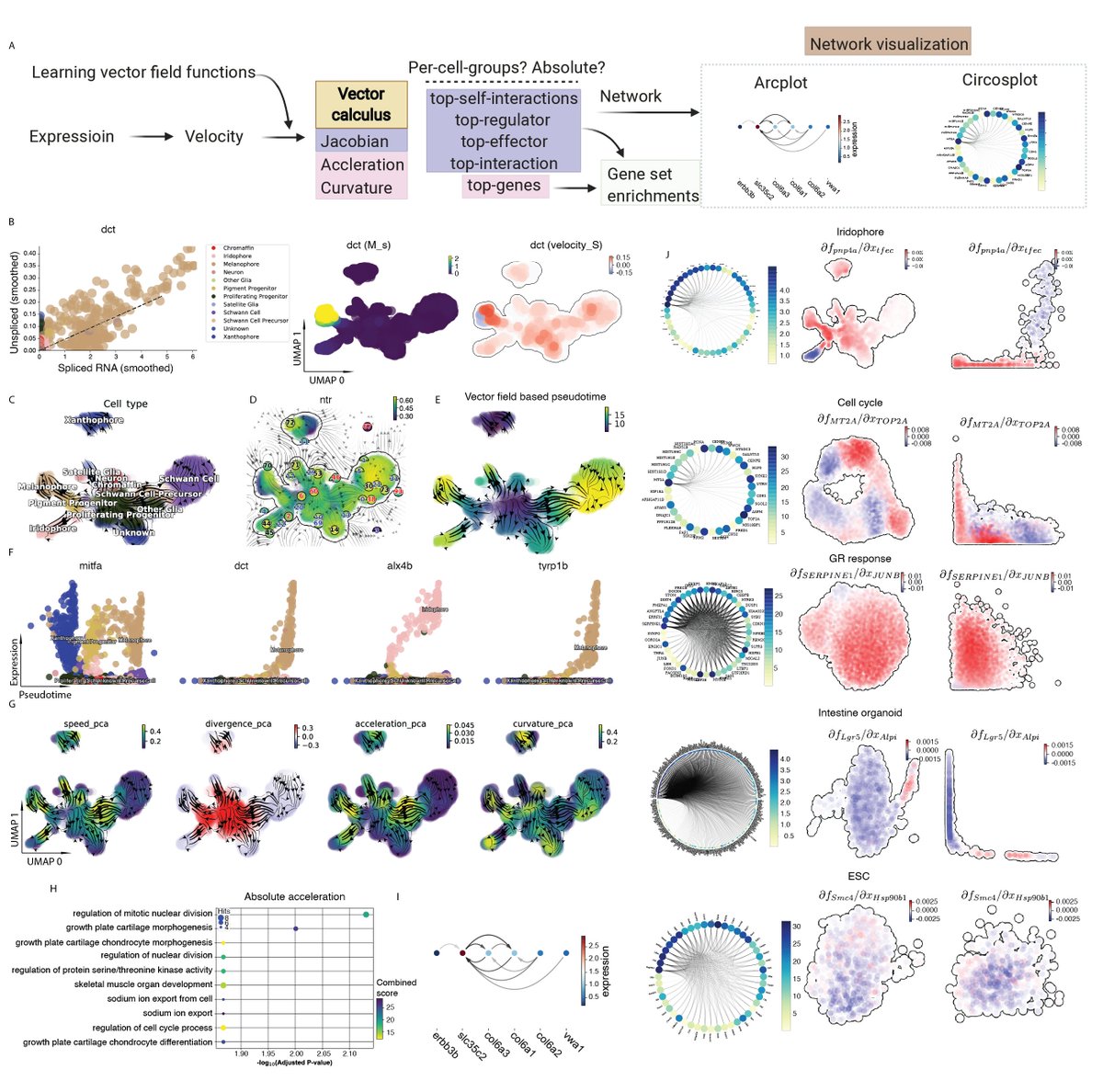
We next move beyond noisy velocity samples to differentiable transcriptomic vector field functions thanks to a nontrivial ML method that learns the function in Hilbert space (RKHS) and derives analytical Jacobian, divergence, acceleration, curvature scalably and accurately.
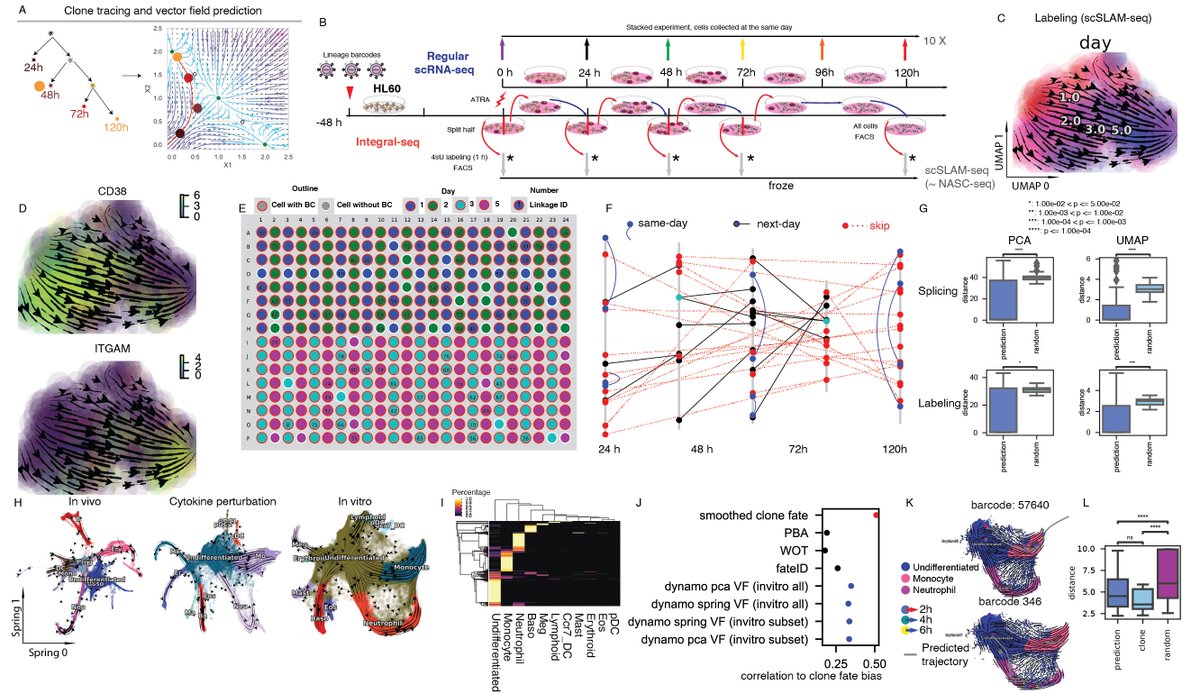
Similar to Newtonian mechanics, if one knows the position of a cell in the state space and our vector field function, one can predict where it is and how fast it is moving, at any point in time. We validated this prediction w/ sequential tracing of single cells of hematopoiesis.
We next applied differential geometric analyses to a variety of conventional or tscRNA-seq data. We reveal the previously unknown zebrafish cell may be derived from chondrocytes, and confirm mutual inhibition between Arx and Parx during iset cell genesis.
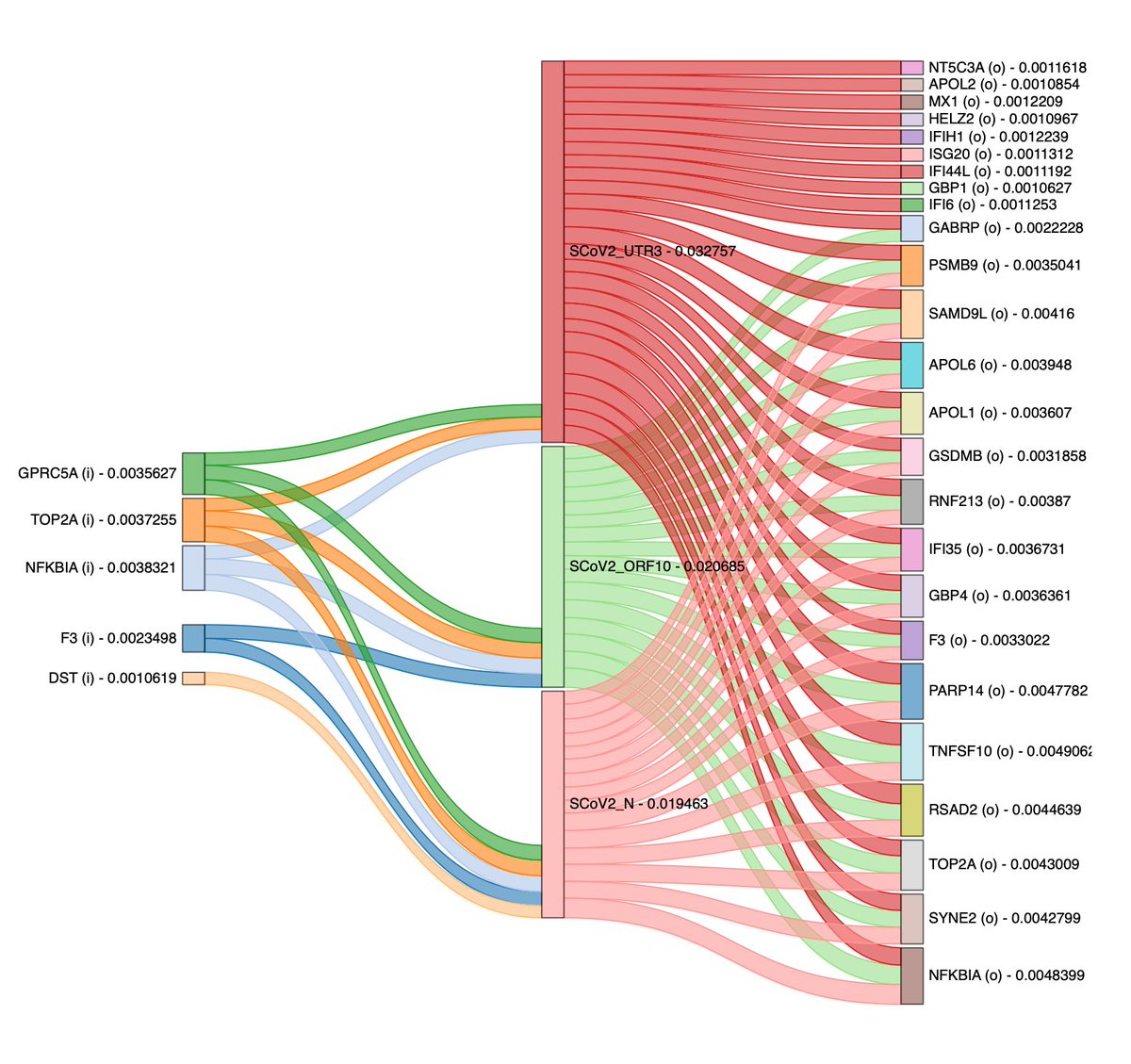
I always hoped my research could contribute to society, e.g providing insights into the pathogenicity of SARS-CoV-2. Composite velocity field analyses of both and virus genes by defining a virus precursor, revealed possible host-virus interaction and resistance mechanisms.
Please also check our Boxes which explains dynamical systems/differential geometry for everyone and provide some intuition for the nontrivial vector field reconstruction algorithm. We believe dynamo is broadly applicable, please reach out to us if you are interested in it.
We all suffered during 2020 but this work was a wonderful journey. I would especially like to thank co-first author @YanZhan92480937 from @jhxing001 Lab. Without our collaboration, this is not possible. I welcome any suggestions/comments, and hope 2021 is a great year for all!

 Read on Twitter
Read on Twitter