My latest paper “Emergence and propagation of epistasis” is out in @eLife. Quite a weight off my shoulders. Some context, results and speculations in this  https://elifesciences.org/articles/60200
https://elifesciences.org/articles/60200
 https://elifesciences.org/articles/60200
https://elifesciences.org/articles/60200
The idea for this project first arose in 2011 after I read Costanzo et al. (Science, 2010). I found Figure S1B particularly interesting. The distribution of epistasis coefficients is perfectly centered at zero. Why? https://science.sciencemag.org/content/327/5964/425.abstract
In this case, it is probably because of measurement noise, but the bigger question is: What distribution should we expect? Same question arose again in my data (Figure 3 in this 2014 paper). Is diminishing returns epistasis surprising or trivial? https://science.sciencemag.org/content/344/6191/1519
The problem is that there is no biologically reasonable null expectation for epistasis. (As opposed to a statistically convenient expectation 𝜀 = 0.) I am aware of Fisher’s geometric model and happy to discuss.
So, how do we go about constructing such null model?
So, how do we go about constructing such null model?
We could compute epistasis in an FBA model. But what would we learn if observed epistasis deviated from the predictions? Most likely, we would learn that the model is deficient. But it probably would not lead us to question whether the data itself are surprising.
For that, we need to know what kinds of epistasis arise generically, i.e., in some large class of systems. My new paper constructs a simple model of a large class of reasonable metabolic networks and asks what types of epistasis arise in such networks.
One property of complex networks in living systems is their hierarchical structure. For example, proteins form pathways. This is critical because epistasis always arises at lower levels and then “propagates” to higher level of biological organization.
For example, two residues in a protein might physically interact, giving rise to epistasis at the level of protein activity. Epistasis for activity can then induce epistasis for fitness, where it will be perceived by natural selection.
So, I ask two questions in this paper. (1) How does epistasis propagate through a metabolic hierarchy? (2) And what kinds of epistasis arise in metabolic networks in the first place?
Turns out, it is possible to derive quite strong results in this simple model.
Turns out, it is possible to derive quite strong results in this simple model.
Result 1. There are three regimes of epistasis propagation. Negative lower-level epistasis always induces negative epistasis at all higher levels, irrespectively of network details. Strong positive lower-level epistasis induces strong positive epistasis at all higher levels.
How weak positive epistasis at a lower level propagates to higher levels depends on the details of the network.
Result 2. What epistasis emerges between two mutations depends on the topological relationship between reactions affected by the mutations.
Result 2. What epistasis emerges between two mutations depends on the topological relationship between reactions affected by the mutations.
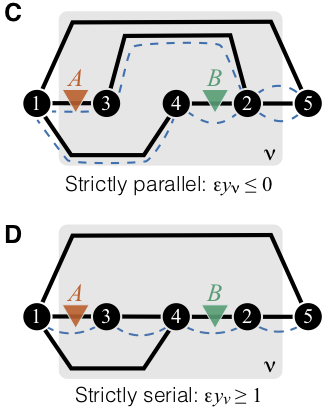
Specifically, if two mutations affect strictly serial reactions, epistasis is strongly positive. If two mutations affect strictly parallel reactions, epistasis is zero or negative. See figure.
These results have some important implications.
 If epistasis arises at some level of the hierarchy, it will not disappear at higher levels. Instead it will induce some epistasis at higher levels.
If epistasis arises at some level of the hierarchy, it will not disappear at higher levels. Instead it will induce some epistasis at higher levels.
 If epistasis arises at some level of the hierarchy, it will not disappear at higher levels. Instead it will induce some epistasis at higher levels.
If epistasis arises at some level of the hierarchy, it will not disappear at higher levels. Instead it will induce some epistasis at higher levels.
 Epistasis for high-level phenotypes, such as fitness, should be ubiquitous. Our default expectation should be that any two mutations that affect fitness are epistatic with each other. Of course, some of these interactions may be weak and therefore undetectable.
Epistasis for high-level phenotypes, such as fitness, should be ubiquitous. Our default expectation should be that any two mutations that affect fitness are epistatic with each other. Of course, some of these interactions may be weak and therefore undetectable.
 Since epistasis depends on network topology, there is no reason to expect that positive and negative epistasis are equally prevalent. For the same reason, there should be many G x G x G interactions (higher-order epistasis) and G x G x E interactions (environmental epistasis).
Since epistasis depends on network topology, there is no reason to expect that positive and negative epistasis are equally prevalent. For the same reason, there should be many G x G x G interactions (higher-order epistasis) and G x G x E interactions (environmental epistasis).
 If the details of the network are unknown, epistasis at lower levels constrains but does not uniquely determine epistasis at higher levels. So, one has to be careful interpreting how measurements of epistasis for, say, enzyme activity translate into epistasis for fitness.
If the details of the network are unknown, epistasis at lower levels constrains but does not uniquely determine epistasis at higher levels. So, one has to be careful interpreting how measurements of epistasis for, say, enzyme activity translate into epistasis for fitness.
 Conversely, epistasis coefficients measured at the level of fitness constrain but do not uniquely specify the underlying functional relationship between gene products. So, we should be careful with the functional interpretations of epistasis measurements.
Conversely, epistasis coefficients measured at the level of fitness constrain but do not uniquely specify the underlying functional relationship between gene products. So, we should be careful with the functional interpretations of epistasis measurements.
As usual, I would love to know your thoughts and/or comments and/or criticisms. Please respond here, DM me, or send me an email.

 Read on Twitter
Read on Twitter