My new preprint, "Why do species get a thin slice of π? Revisiting Lewontin’s Paradox of Variation" is up on BioRxiv! Now that I’ve had  , I’ll step through some of the major points in this paper. Link: https://www.biorxiv.org/content/10.1101/2021.02.03.429633v1
, I’ll step through some of the major points in this paper. Link: https://www.biorxiv.org/content/10.1101/2021.02.03.429633v1
 , I’ll step through some of the major points in this paper. Link: https://www.biorxiv.org/content/10.1101/2021.02.03.429633v1
, I’ll step through some of the major points in this paper. Link: https://www.biorxiv.org/content/10.1101/2021.02.03.429633v1
Lewontin’s Paradox of Variation is an old mystery: if neutral theory says that genetic diversity (i.e., π) should grow with population size (π ≈ 4Nμ), and we know census population sizes (Nc) vary over several orders of magnitude, why is the range of π across species so narrow?
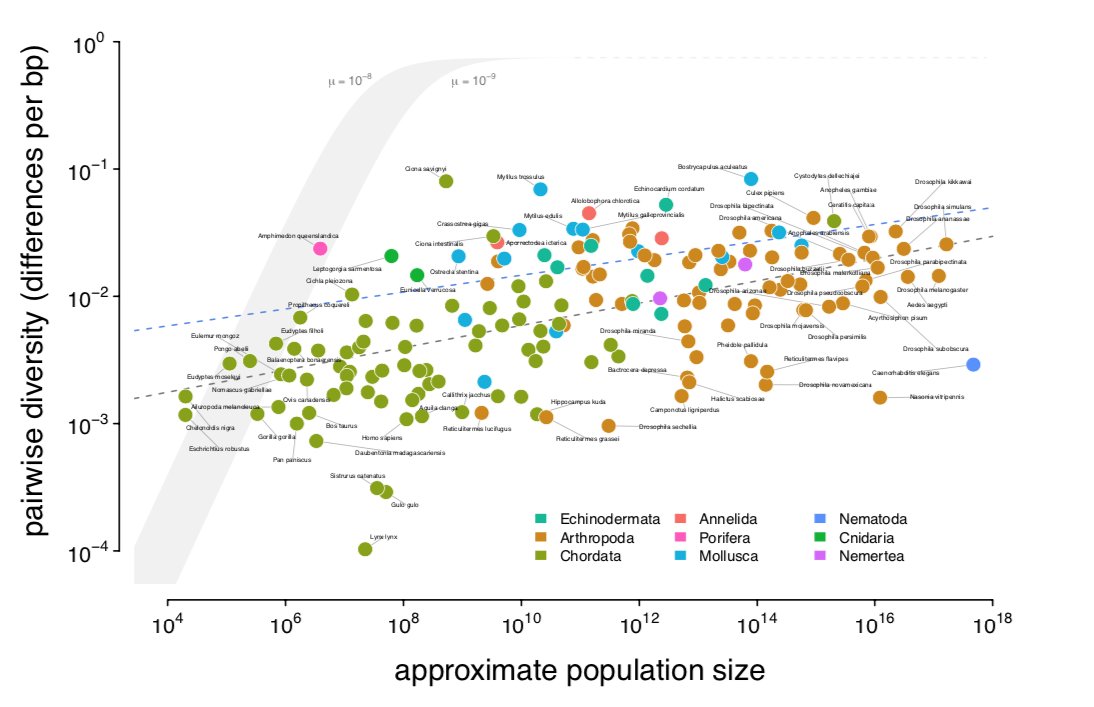
First, inspired by some great past surveys of the heterozygosity–census size relationship (Soulé '76, Nei & Graur '84) I wanted to estimate this relationship using genomic estimates of π. But census size is hard to estimate; my approach was to approximate, Nc ≈ range x density.
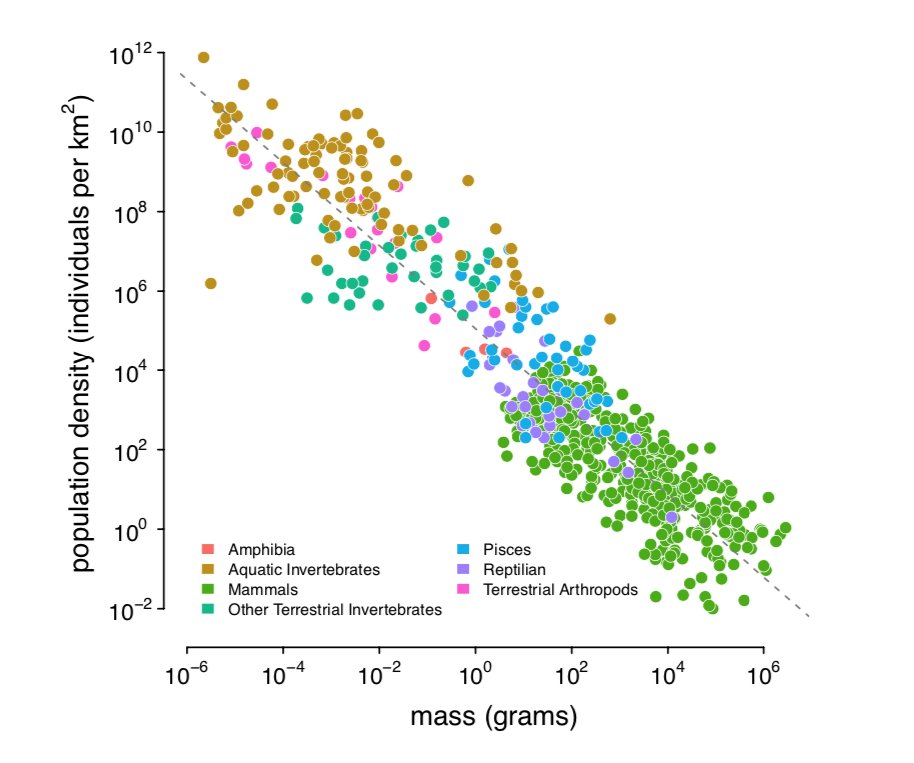
Damuth ('87) has shown that across metazoans there's a beautiful relationship between pop density and body size (right) which I combine with ranges I estimated from occurrence data for 172 species. With π estimates from @ellenleffler's and other surveys, I get (left):
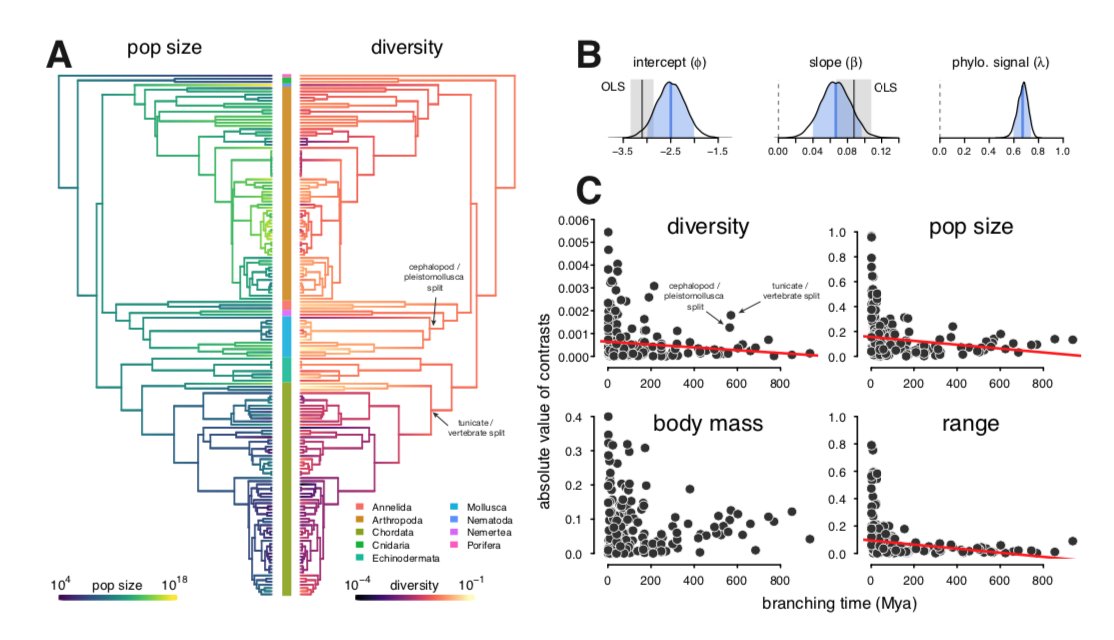
I then was curious whether this across-species relationship was significant accounting for phylogeny. Generally, PCMs have not been widely used in across-taxa popgen ( ). Using phylo mixed-effects models, I find it is significant (B) with some other interesting things going on.
). Using phylo mixed-effects models, I find it is significant (B) with some other interesting things going on.
 ). Using phylo mixed-effects models, I find it is significant (B) with some other interesting things going on.
). Using phylo mixed-effects models, I find it is significant (B) with some other interesting things going on.
Some have argued that since coalescent times are << divergence times, we don't need to worry about PCMs for π. I find high phylo signal (B above) and using node-height tests, deep rate shifts in π that suggest this is incorrect (see discussion). Basically, PCMs in popgen =  .
.
 .
.
Finally, selection. Could selection explain this? There's a nice history of work on this, e.g. @RussCorbett et al. ('15) and @Graham_Coop ('16). With my Nc estimates, I was curious: if we do the extreme thing of assuming Ne = Nc, do recurrent hitchhiking and BGS models even work?
But we lack parameter estimates for most species. Well, what if we use *really* strong sel estimates from Drosophila? Gives linked selection the best chance. Then, I consider a major determinant of linked selection: recombination (using the  dataset from @jessstapley et al.).
dataset from @jessstapley et al.).
 dataset from @jessstapley et al.).
dataset from @jessstapley et al.).
Fascinatingly, big Nc species have little recombination (linked sel  , see A)! With these really, really high parameter estimates, we can make predicted π ≈ observed, but mid-Nc species still don't fit because of their long map lengths (linked sel
, see A)! With these really, really high parameter estimates, we can make predicted π ≈ observed, but mid-Nc species still don't fit because of their long map lengths (linked sel  , B).
, B).
 , see A)! With these really, really high parameter estimates, we can make predicted π ≈ observed, but mid-Nc species still don't fit because of their long map lengths (linked sel
, see A)! With these really, really high parameter estimates, we can make predicted π ≈ observed, but mid-Nc species still don't fit because of their long map lengths (linked sel  , B).
, B).
So the idea that these models of linked selection could reduce π from levels implied by census sizes is a little suspect. Also, for mid-Nc species π to be reduced to observed levels, the selection parameters would have to be higher than in Drosophila 
 .
.

 .
.
(and even to bring π down for  given Nc, current π0 estimates would have to be way off
given Nc, current π0 estimates would have to be way off  !).
!).
These are all simple selection models though... lots of discussion in, well, the Discussion about what could be going on here. Also lots more I didn't include. Thanks for following!
 given Nc, current π0 estimates would have to be way off
given Nc, current π0 estimates would have to be way off  !).
!). These are all simple selection models though... lots of discussion in, well, the Discussion about what could be going on here. Also lots more I didn't include. Thanks for following!
By the way, happy to receive feedback! Also, this project was entirely from available data and used a ton of great open source packages (i.e. the *super cool* datelife pkg from @omearabrian, @LunaSare et al!).

 Read on Twitter
Read on Twitter