Our report on the genomic characterisation of the first wave of the SARS-CoV-2 epidemic in South-Africa is now out in Nature Medicine, a study which was crucial to confidently describe the 501Y.V2 variant which emerged after this time-frame. [THREAD]
https://www.nature.com/articles/s41591-021-01255-3
https://www.nature.com/articles/s41591-021-01255-3
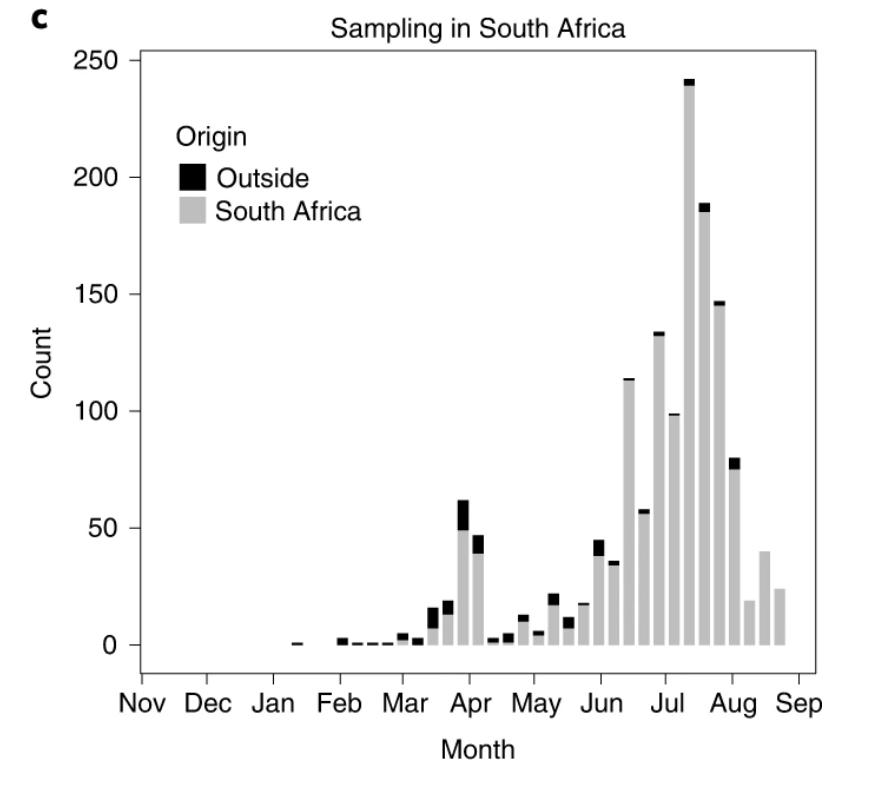
This is the result of consistent genomic surveillance from the beginning of the country’s epidemic in March and more extensively during the peak of infections, thanks to a great collaboration of public and private institutions within the Network for Genomic Surveillance in SA.
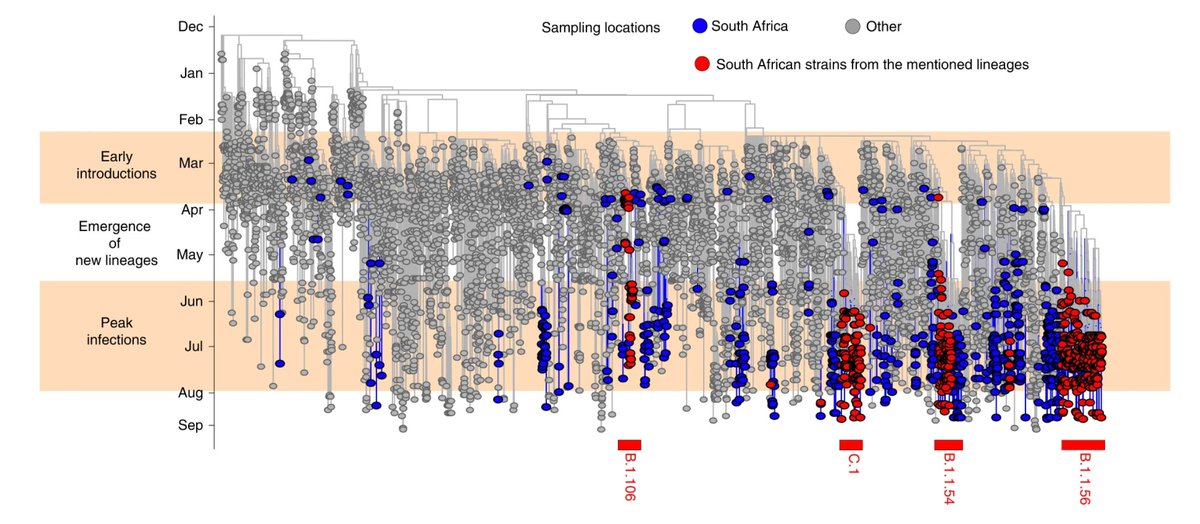
Our analysis of 1365 genomes in this study shows that the bulk of introductions happened in March/beginning of April and that most infections following this were local transmissions.
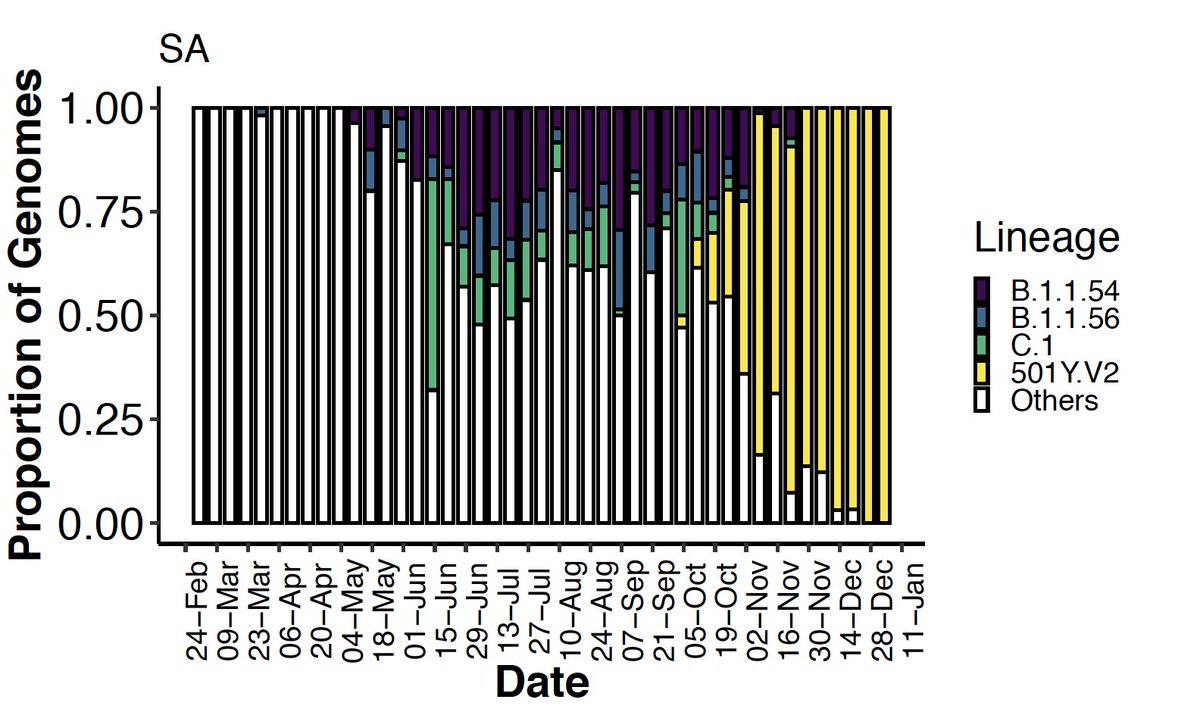
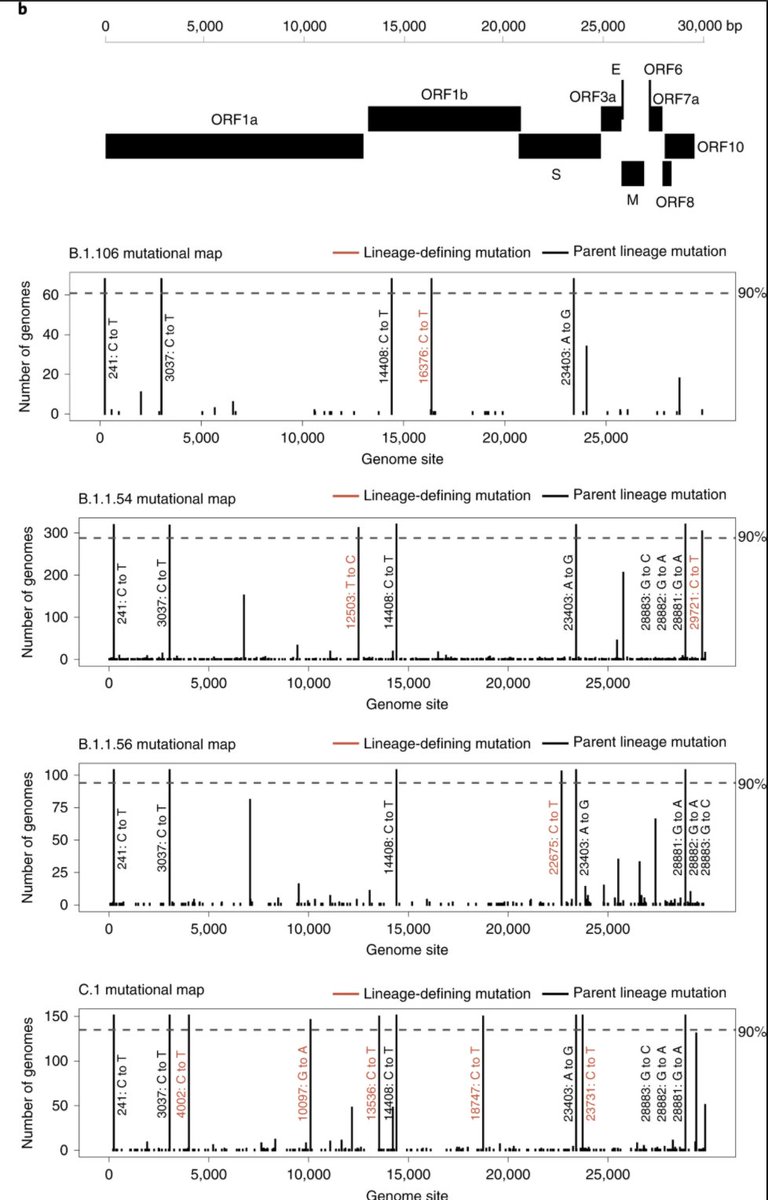
We identified 16 lineages specific to South Africa that emerged during lockdown and circulated widely during the first wave. We focus on 4 main ones, labelled in red below: B.1.106, B.1.1.54, B.1.1.56 and C.1.
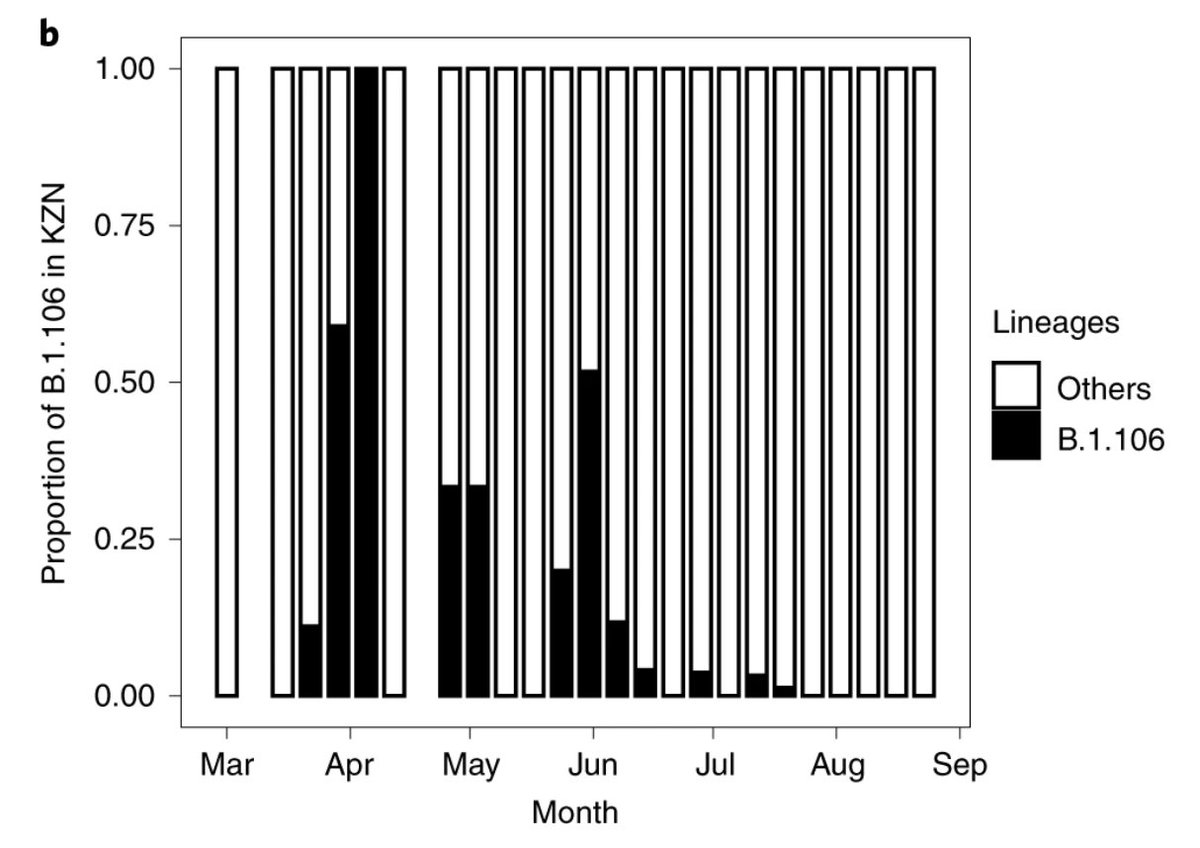
B.1.106 was an early circulating lineage in the province of KZN following amplification in nosocomial outbreaks in March, at one point accounting for 100% of the infections in the province, and completely disappearing following genomic-informed outbreak control measures.
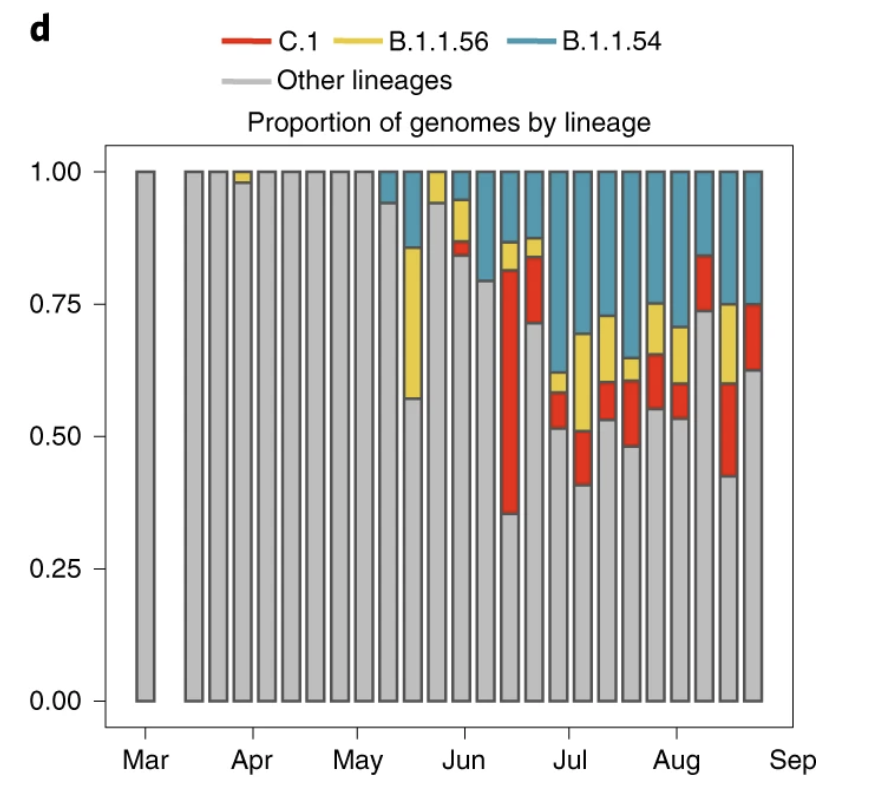
Lineages B.1.1.54, B.1.1.56 and C.1 spread widely in South Africa during the first wave, comprising ~42% of all infections in the country at the time. A jump in the future to January clearly shows the overwhelming replacement of these lineages by 501Y.V2.
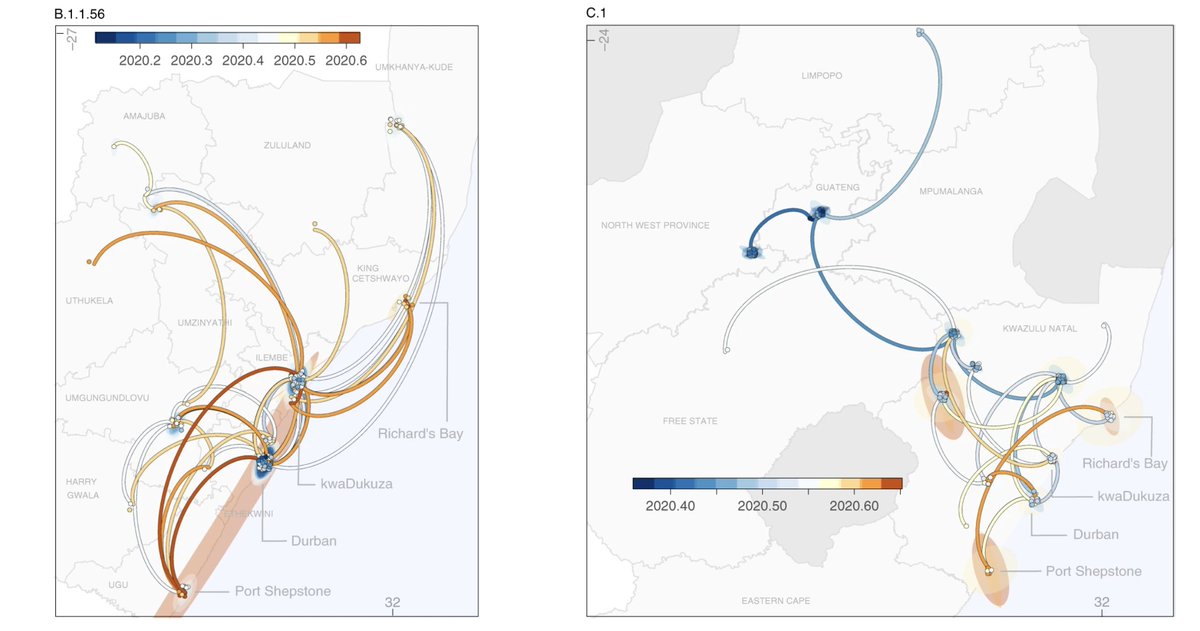
Phylogeographic analysis revealed that B.1.1.56 originated in the city of Durban and spread widely in the province of KZN while the C.1 lineage originated in Gauteng and spread to 4 provinces and later also identified in the city of Cape Town and neighbouring Mozambique.
Despite widespread circulation in the country, the main lineages described in this study only harbor D614G amino acid mutation in the Spike region, suggesting no fitness advantage and increased transmission merely due to the epidemic dynamics of the country.
This is in stark contrast with the later 501Y.V2 with 7-8 amino acid changes in Spike and clear fitness advantage. Without such a thorough genomic surveillance program from the very first infections in South Africa, we would not have been confident to characterise the new variant
This work and all ongoing work are an incredible collaboration between the @krisp_news team and beyond and would not have been possible without hard-work from many in the labs, in the clinics and behind computers. @Tuliodna @EduanWilkinson @rjlessells @Mittenavoig @JSAN4CHRIST
Most importantly, it shows how crucial sequencing and genomic surveillance are in this fast-evolving pandemic. When we wrote this paper, we had no idea of what was coming in the months that followed.

 Read on Twitter
Read on Twitter![Our report on the genomic characterisation of the first wave of the SARS-CoV-2 epidemic in South-Africa is now out in Nature Medicine, a study which was crucial to confidently describe the 501Y.V2 variant which emerged after this time-frame. [THREAD] https://www.nature.com/articles/s41591-021-01255-3 Our report on the genomic characterisation of the first wave of the SARS-CoV-2 epidemic in South-Africa is now out in Nature Medicine, a study which was crucial to confidently describe the 501Y.V2 variant which emerged after this time-frame. [THREAD] https://www.nature.com/articles/s41591-021-01255-3](https://pbs.twimg.com/media/EtN0L4WVcAEZxED.png)