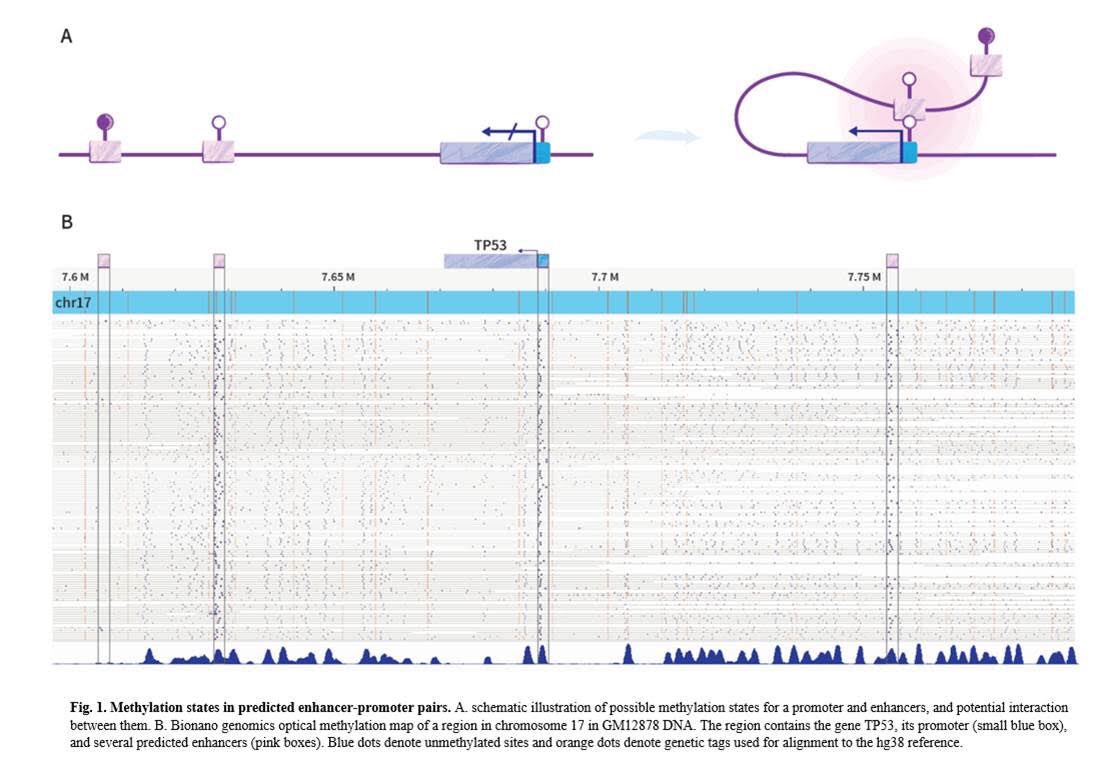
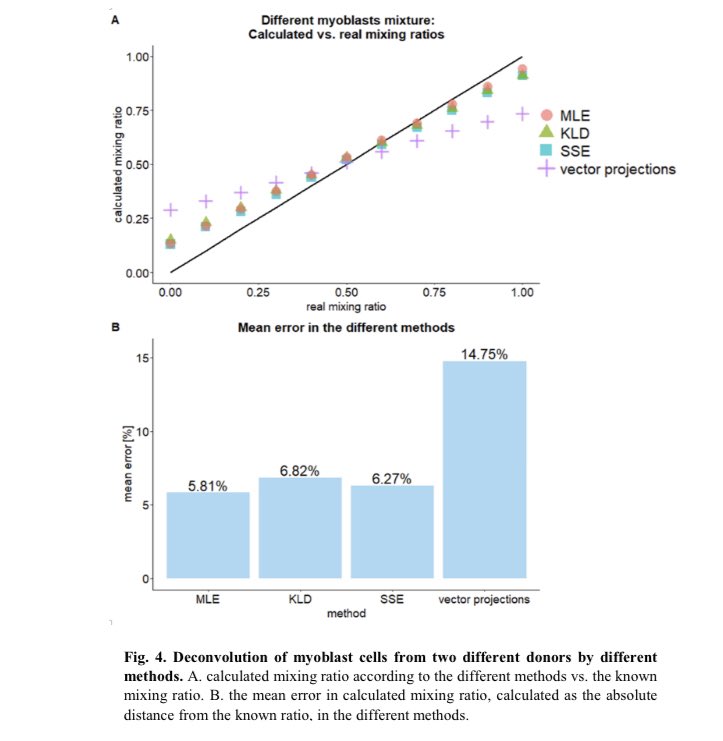
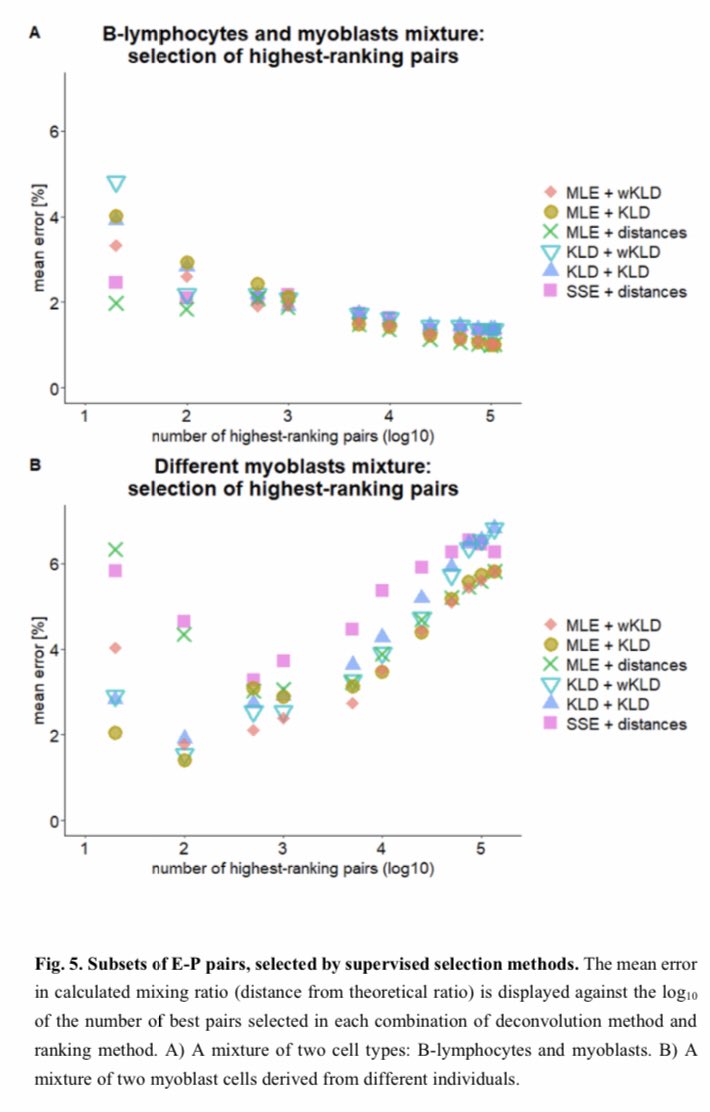
I think it’s the first time to be done...analyzing methylation of promoters and distant enhancers on the same long DNA molecule, millions of long reads, over 100,000 E-P pairs @ 30x-300x with @bionanogenomics optical methylation mapping. 1/3 https://www.biorxiv.org/content/10.1101/2021.01.28.428654v1
This is the computational framework for using this new type of data for cell mixture deconvolution for cell mixtures with similar genetic/epigenetic background like in cancer, where enhancers cause more methylation variance than promoters

 Read on Twitter
Read on Twitter