1/14 A Friday arvo summary of our @DETECTIVE_AMR manuscript that describes a novel group of plasmids carrying antibiotic resistance genes in Acinetobacter: https://pubmed.ncbi.nlm.nih.gov/33501980/
2/14 Most isolates in our collection of carbapenem-resistant A. baumannii isolated in a Hangzhou ICU belong to global clone 2 and produce OXA-23. I find our rare non-GC2 isolates with different carbapenem resistance genes more interesting.
3/14 Enter isolate DETAB-P2 from a patient rectal swab. It’s unusual - ST138 by the Pasteur scheme and a novel type, ST2209 by the Oxford scheme. It carries genes for OXA-58 and NDM-1.
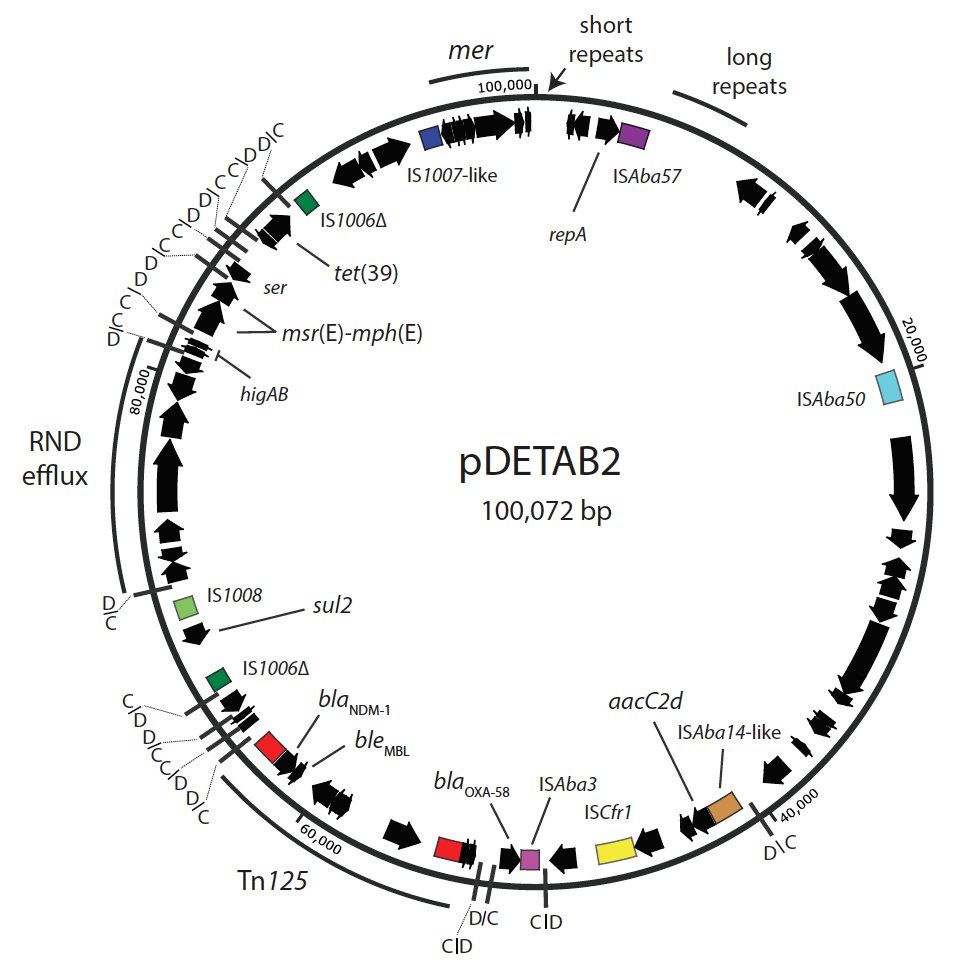
4/14 A hybrid genome assembly yielded the 100,072 bp plasmid pDETAB2. It contains the carbapenamase genes plus genes conferring resistance to aminoglycosides, tetracycline, sulphonamides and macrolides. Most interestingly, there are 16 pdif sites in pDETAB2.
5/14 What are pdif sites? They’re recognised by XerC/XerD recombinases, and in Acinetobacter plasmids they flank dif modules that appear to move between sites - @GraceBlackwel1 and Ruth’s manuscript provides a great introduction! https://pubmed.ncbi.nlm.nih.gov/28533235/
6/14 ll of the resistance genes in pDETAB2 are inside dif modules. Even the NDM-1 gene is in a complete Tn125 that’s inserted into a tiny dif module containing putative toxin-antitoxin genes.
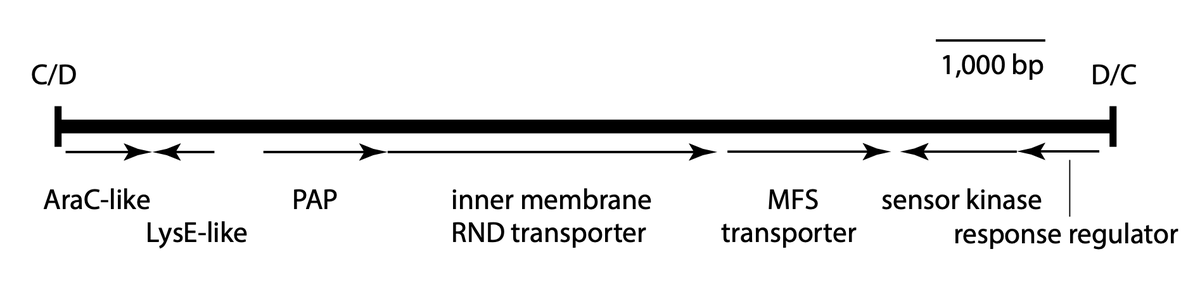
7/14 dif modules don’t just carry resistance genes! An interesting one in pDETAB2 appears to encode a novel RND efflux pump - we were very lucky to have @JessicaMABlair run an expert eye over it for us to confirm!
8/14 pDETAB2 is a new replicon type, GR34. When we queried GenBank with pDETAB2’s repA, we found identical genes in 18 plasmids. They’re from various Acinetobacter species from diverse environments - my favourite being an A. ursingii isolated from duckweed in Japan.
9/14 Comparing these plasmids wasn’t easy! GR34 plasmids don’t have a ‘backbone’ like plasmids of E. coli do. Instead, they share only a 10 kbp region that includes the rep gene, putative partitioning + toxin-antitoxin genes and some funky repeats.
10/14 Apart from that region, these plasmids are mostly made of dif modules. All sorts of dif modules, many encoding hypothetical proteins. Some are shared by plasmids from different species and environments, but even those appear in different combinations and orientations.
11/14 GR34 plasmids are mercurial mosaics. This successful replication platform is associated with all sorts of accessory content. I’d love to know where module exchange is occurring and how quickly a plasmid’s module content changes when it enters a new host/environment.
12/14 Acinetobacter plasmids are weird! GR34 plasmids and their dif modules might be important contributors to the dissemination of resistance determinants in the wider Acinetobacter genus, and I’m sure we’ve plenty more to learn about them.
13/14 For some more exciting dif module discussion, check out this very recent paper from @MoHamidian, @GraceBlackwel1, @DrSNigro, Steph and Ruth (I haven't had the chance to read it properly yet myself!) https://pubmed.ncbi.nlm.nih.gov/33452522/
14/14 If you’ve made it this far, you’re a legend. Thanks to Haiyang Liu, @Emma_L_Doughty, @XiaotingHua, @alanmcn1, @WvSchaik and other @DETECTIVE_AMR collaborators. More exciting stories to come!

 Read on Twitter
Read on Twitter