1/7 This article reflects how scientific collaboration is working across international borders with online colleagues that are willing to share their expertise openly. Using the UK's genomic early warning system @CovidGenomicsUK we spotted a mutation that concerned us - N439K https://twitter.com/CellCellPress/status/1354777858351181826
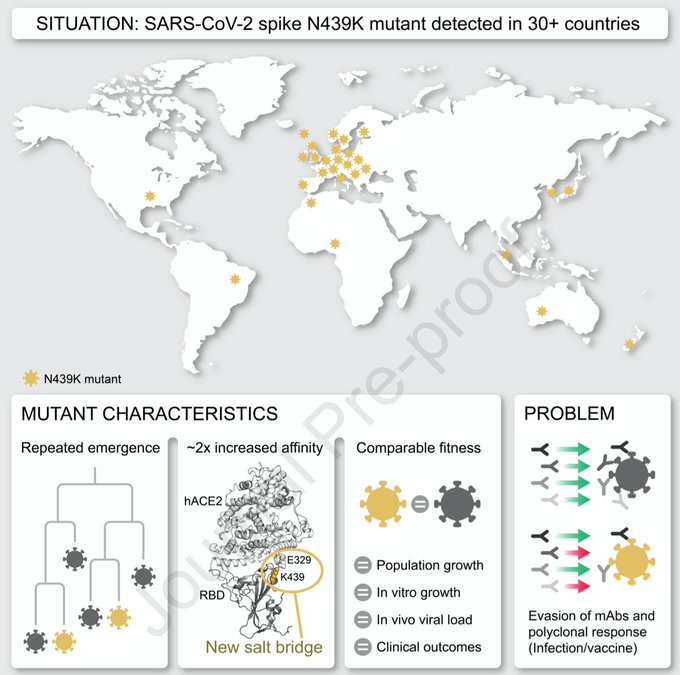
2/7 This change from asparagine to lysine at site 439 in spike (first detected in Scotland @NHSGGC ) looked like it might increase binding to the human ACE2 receptor. Our international colleagues confirmed that binding affinity was increased by the formation of a new salt bridge
3/7 Neutralisation was also affected with reduced monoclonal and polyclonal antibody responses (including one of the Regeneron antibodies). Chris Davis ran cross-competition assays in our high containment labs and found that the variant was not attenuated by this change.
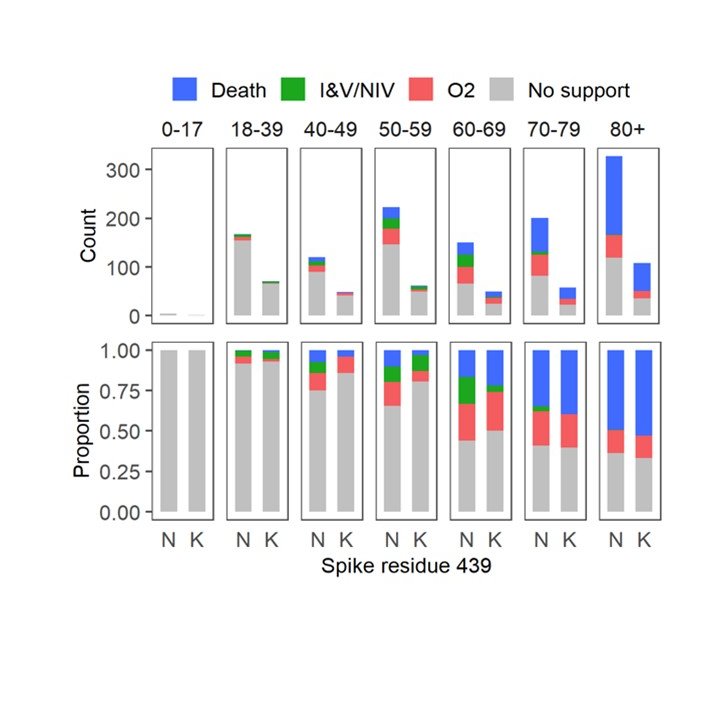
4/7 439K was associated with higher viral loads. Ruth Jarrett @CVRinfo set up RT-PCR testing using RNA standards to quantify this. Our clinical fellow team including @JG_Shepherd @ngjesudason @DrLi_Virology @Rajiv_334 showed that clinical outcomes were similar.
5/7 The sequencing and analysis was funded by the COG-UK consortium and led by @CVR_Genomics and @robertson_lab who are working night and day to monitor virus evolution across Scottish health boards alongside our colleagues in NHS Lothian and the rest of the UK.
6/7 The variant that arose in Scotland became extinct following the first national lockdown - but the N439K change arose again independently in Europe and is now one of the more common changes seen in the genome in 30+ countries.
7/7 We now know that spike can accommodate multiple changes as seen in new variants. We expect to see selection in the genome following vaccination. Continued monitoring supported by @CovidGenomicsUK and open data sharing will help vaccine developers to update these as needed.

 Read on Twitter
Read on Twitter