. @BrieucLehmann from @oxcsml @OxfordStats, with @cholmesuk & Gil McVean just released a pre-print @biorxiv_genomic (which I meagrely contributed to & y'all should know about) on the transferability of polygenic scores across different ethnicities
https://www.biorxiv.org/content/10.1101/2021.01.15.426781v1


https://www.biorxiv.org/content/10.1101/2021.01.15.426781v1


For non- #genetics folks, a polygenic score (PGS) is a measure of how much a certain trait is determined by a genetic factor. Like with much of medical sciences, many of the PGS have been developed on predominantly white populations. This is bad.
Individuals from different ethnic groups will tend to have different ancestral backgrounds which have some genetic differences(there are of course many other contributing factors!). PGS trained on predominantly white/European samples tend to perform badly in other groups
See?Bad
See?Bad
The solution is ultimately to increase the representation of under-represented ethnicities/ancestries in genetic datasets but this takes a long time and is expensive.
So can a wee bit of statistics using the data we have help in the interim?
So can a wee bit of statistics using the data we have help in the interim?
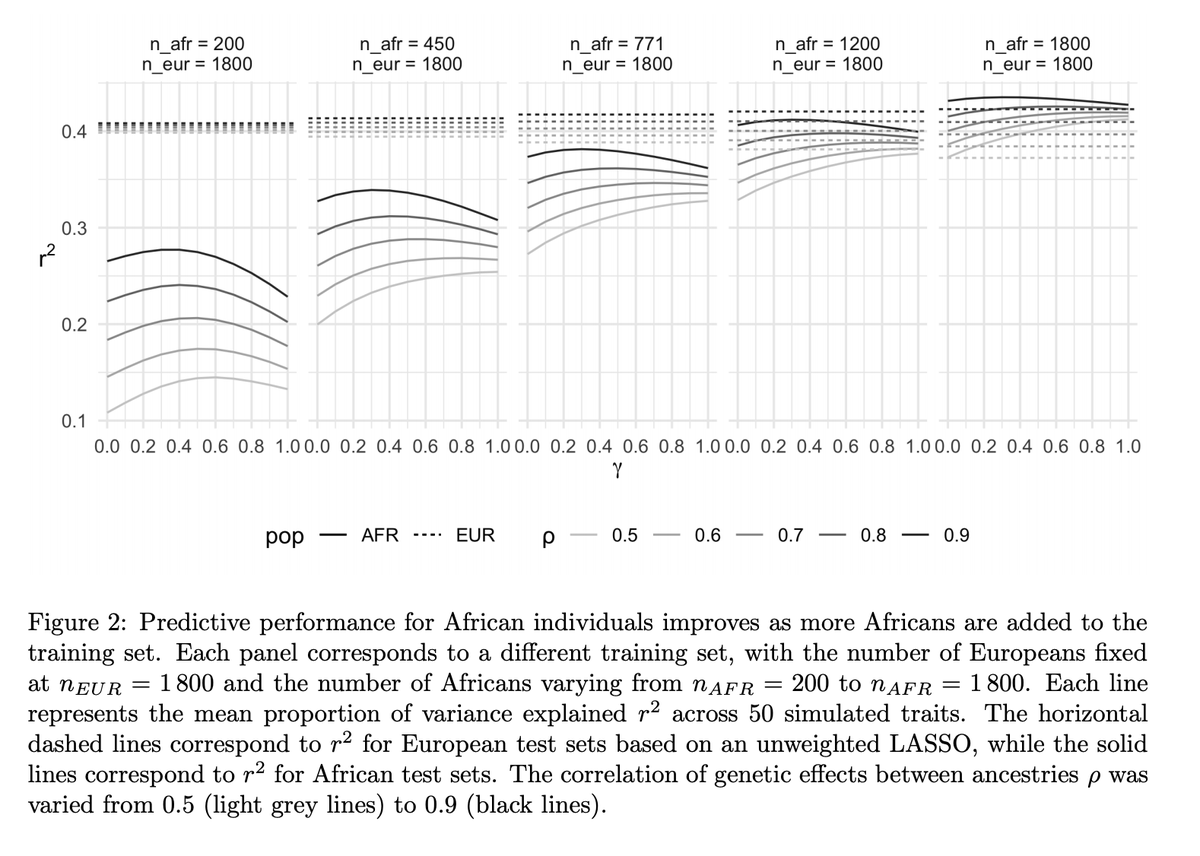
So, we looked at how multiple-ancestry training sets (e.g. in @uk_biobank) can be used to improve PGS for individuals from underrepresented groups.
Here are some highlights:
Here are some highlights:
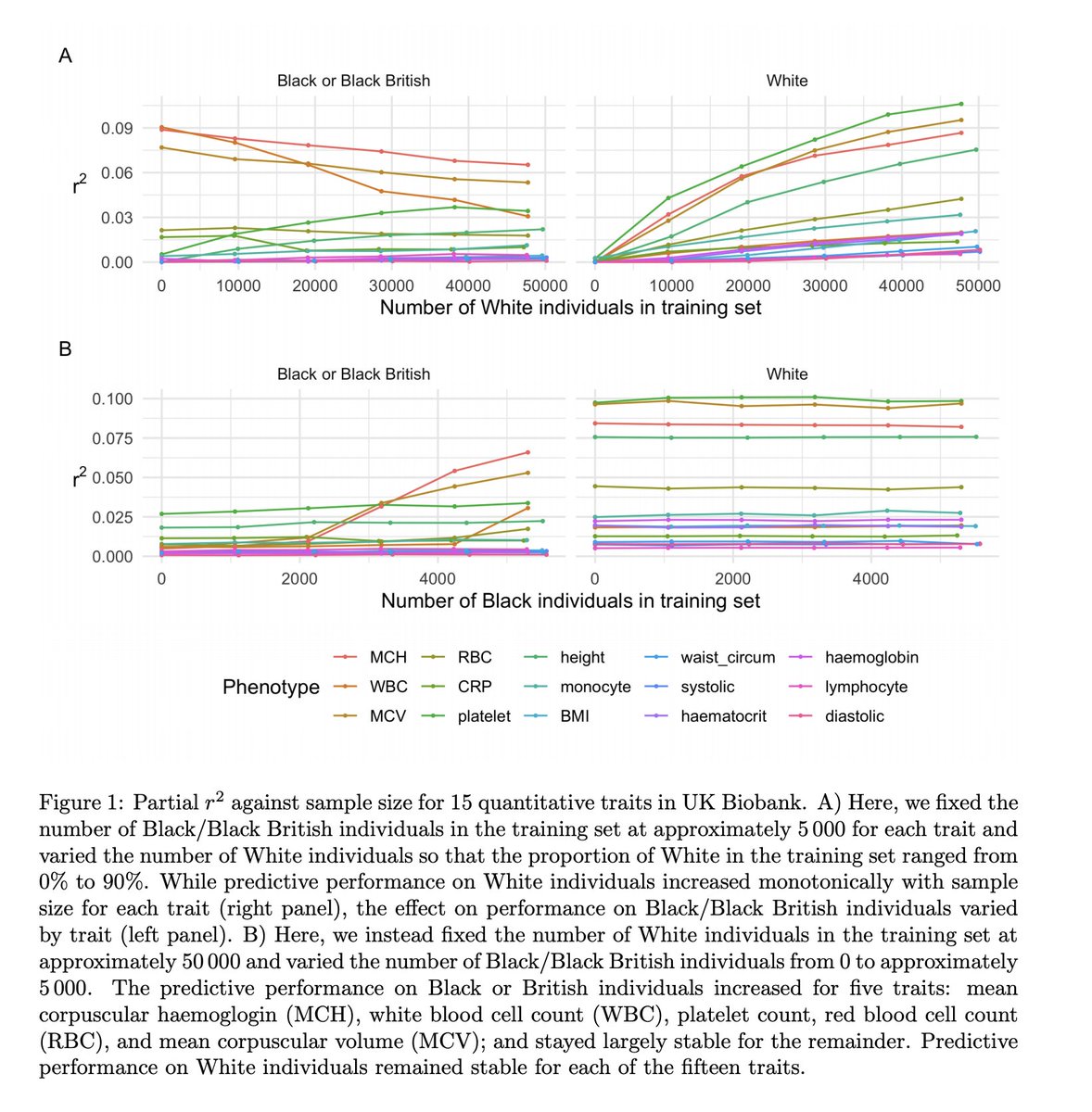
 Headline 1: Adding individuals from one ancestry does not always improve PGS performance for a different ancestry
Headline 1: Adding individuals from one ancestry does not always improve PGS performance for a different ancestry
We used training sets with a varying numbers of White & Black ppl. For SOME of the 15 traits, adding more White ppl resulted in worse performance for Black ppl
 Headline 2: Importance re-weighting can provide modest improvement in PGS performance
Headline 2: Importance re-weighting can provide modest improvement in PGS performance
We used importance re-weighting (+ weight to ppl from under-represented groups) to address the ethnicity imbalance artificially. Kiiiiinda worked, but not when the imbalance was big.
Booo
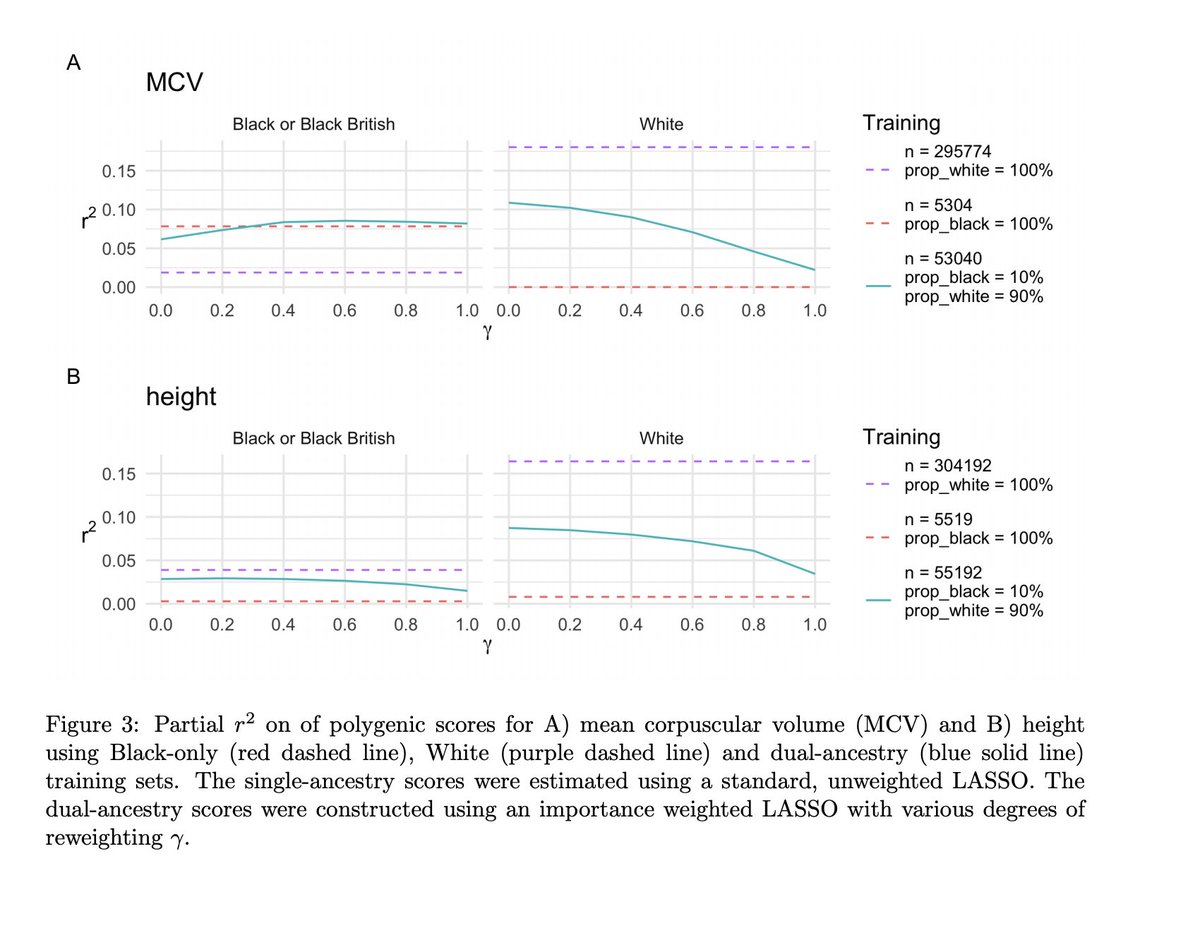
 Headline 3: Optimal ancestry composition of training sets varies among traits
Headline 3: Optimal ancestry composition of training sets varies among traits
Importance re-weighting was v trait-dependent (also in a "good" way!)eg. mean corpuscular volume: PGS trained on a small no of Black ppl far outperformed one from a much larger number of White ppl
 Headline 3b: Differences in trait architecture explain variable performance by ancestry
Headline 3b: Differences in trait architecture explain variable performance by ancestry
We wanted to know WHY optimal training approaches varied across traits by investigating the contribution of variants at different allele frequencies to prediction accuracy
Some things to mull over as you scroll this on the  :
:
- Our White genetic datasets raise many technical, clinical and ethical issues. These will and are impacting on health inequalities
- Large biobanks mean we can investigate artificial solutions to the low n of non-white ppl
 :
:- Our White genetic datasets raise many technical, clinical and ethical issues. These will and are impacting on health inequalities
- Large biobanks mean we can investigate artificial solutions to the low n of non-white ppl
- Based on our re-weighting approach, it's not really good enough to overcome the low number of non-white ppl in PGS
- But a statistical plaster merely exposes the fact there is a whole pipeline where bias and exclusion enter and therefore should be considered in combination
- But a statistical plaster merely exposes the fact there is a whole pipeline where bias and exclusion enter and therefore should be considered in combination
So, COLLECT MORE GENETIC DATA ON NON-WHITE PEOPLE
In doing so, we can therefore also move away from the unhelpful discrete ethnic/ancestry/race boundaries towards continuous representations of genetic ancestry
(hat tip @GenomicsEngland @H3Africa @AllofUsResearch + others!)
In doing so, we can therefore also move away from the unhelpful discrete ethnic/ancestry/race boundaries towards continuous representations of genetic ancestry
(hat tip @GenomicsEngland @H3Africa @AllofUsResearch + others!)
"Ultimately,approaches to genetic prediction must acknowledge both the many similarities of human biology,but also the differences in history,cultural heritage, exposure,& behaviour that can lead to certain factors being of greater relevance for particular groups of individuals"
It's a pre-print yeah, so you still have plenty of opportunity to email me with all your strong opinions weakly held (mmackintosh[at]turing[dot]ac[dot]uk)
May be of interest/you've been reffed @chris_wigley @nicolablackwood @Patient_Data @natalie_banner @irenetrampoline @MarzyehGhassemi @sairaghafur @turinginst @HealthFdn @One_HealthTech

 Read on Twitter
Read on Twitter