For all of you PCR nerds out there, the lab has just sent me over some screenshots of a great example of an N gene variant. That is a virus with a change somewhere along the part of the genome that codes for nucleocapsid, that directly affects probably one of the primers.
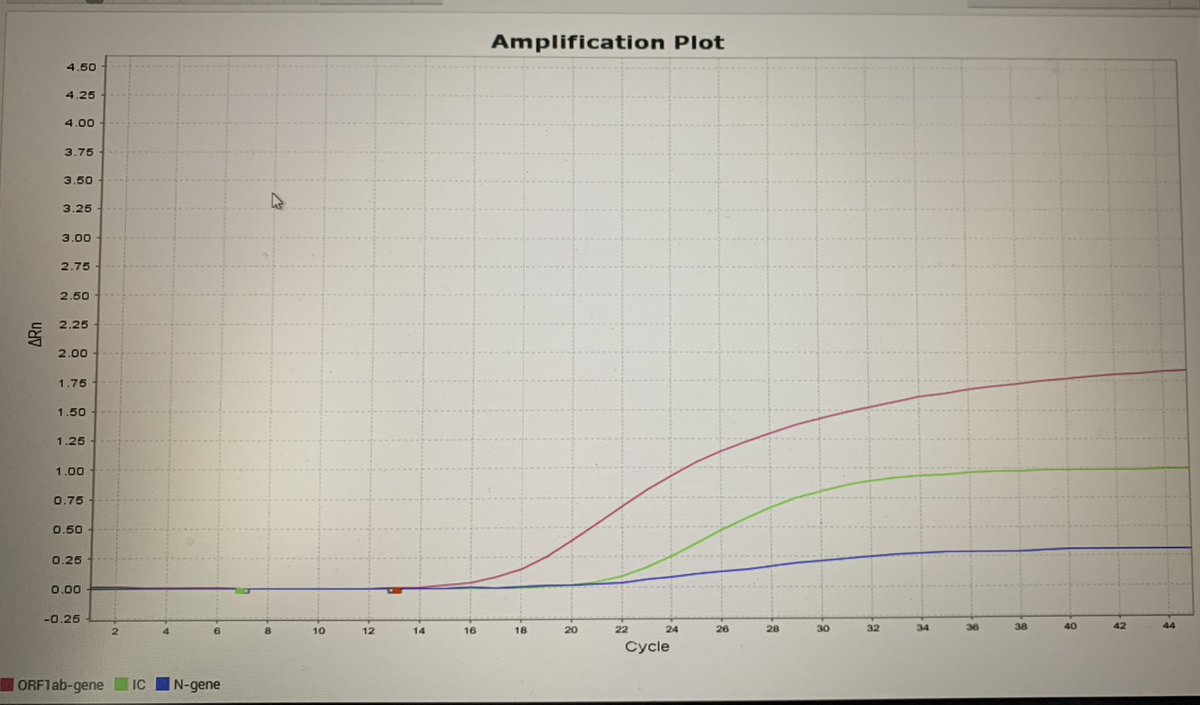
This particular PCR test is a multiplex in a single tube that amplifies the ORF and N of the virus and a human house keeper gene. The amplification curves are red for ORF, green for housekeeper gene and blue for N.
What you see is the ‘linear curve’ for the assay. A PCR has a lag phase (the flat bit where the reaction is happening but the target product isn’t high enough to read), a linear phase (as the target product increases exponentially with the reaction) and a plateaux phase.
What has drawn the attention of the lab (and hence a request for my input) is that the N gene isn’t showing what we would describe as a typical curve, (in fairness the green line is probably the best shape of the three curves as at least the linear phase is sort of straight :)
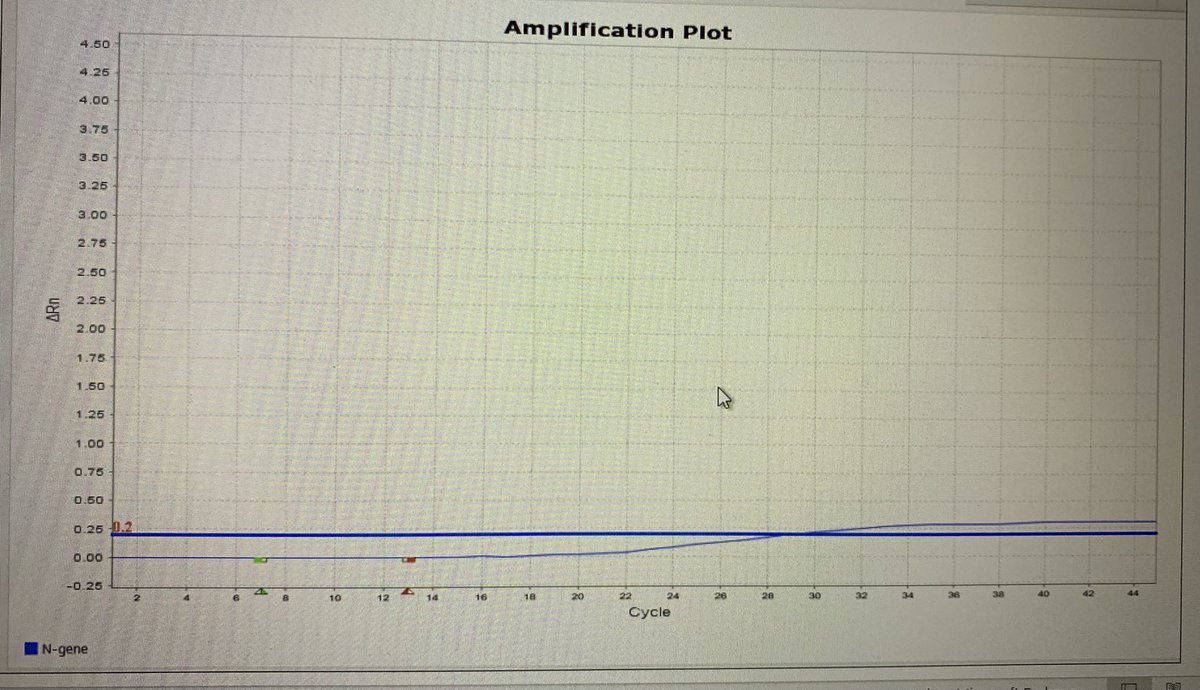
It’s much flatter, so they’ve helped me along by adding in the standard threshold-the point at which the product passes to become positive
You can see from this a shallow linear phase, with the plateaux skimming the normal threshold level. To determine the ct value- draw a line from the point the two blue lines meet downwards- and you’ll meet the number 30. Using that threshold- that’s the Ct value of the N gene.
But it’s wrong. We set a threshold based on the point that the linear phase moves into the exponential growth, so for this target, the threshold is likely to be closer to 1. If you move it, the ct value for N then becomes between 20- 23.
This is actually then much closer to what would be reported for ORF - ct 19. This person is not only positive, but has lots of virus.
So what is going on?
The S gene target failure as described for the UK variant is an example of a virus with a mutation that directly impacts the tests we use. What I’m showing you in this thread is another type affecting a different target (N). I can also show you a virus with one affecting ORF.
So far I’m unable to show you one that affects multiple targets, but I’m sure that will happen given time.
Reassuringly, thanks to the skill of the lab staff in my department running this test today. It was noted and referred for advice quickly. This person will get the positive result they need for management and we’ll have a virus that we can sequence to find what the problem is.
This isn’t unusual in molecular diagnostics, it’s something we normally monitor for. The difficulty for us comes when assays are commercial and the sequences used for the test are proprietary- we have to rely on the companies to monitor for these changes.
In this case, although technically the sensitivity hasn’t been affected- if you move the threshold-it has, if the threshold is fixed by the company. A sample with a lower viral load, could easily be called negative or >35 - when in fact it’s a good going, acute infection.

 Read on Twitter
Read on Twitter