I’m delighted to share our paper ( @nprovine @TheLucyGarner & I) examining human intestinal tissue-resident memory T cells (Trms) in the model of intestinal transplantation. Here's a short thread to describe the findings for those who are interested! https://www.cell.com/cell-reports/fulltext/S2211-1247(20)31650-8#.YArU8CL0q8k.twitter
So, Trm cells are super-cool. These long-lived memory populations sit in all sorts of tissues, particularly mucosal sites. They act as sentinels, ready for rapid activation, including pro-inflammatory and cytotoxic action. They're the crack troops of the adaptive immune response.
Trm cells have interesting roles in human disease, including infections (especially viral), cancer , transplantation, and autoimmunity. Take a look at this ace review from my friend @sarah_sasson to see the full diversity of Trm roles in human disease. https://www.nature.com/articles/s41423-019-0359-1
But Trms are tricky to study in humans, because Trms are defined functionally, by their lack of recirculation. This can be studied in mice more easily (using IV labelling or parabiosis) but it is hard in humans.
Instead we use proxy markers - CD69 and CD103. But these aren’t perfect - CD69 is not specific for Trms, and CD103 is not sensitive (particularly in some tissues, and for CD4 T cells).
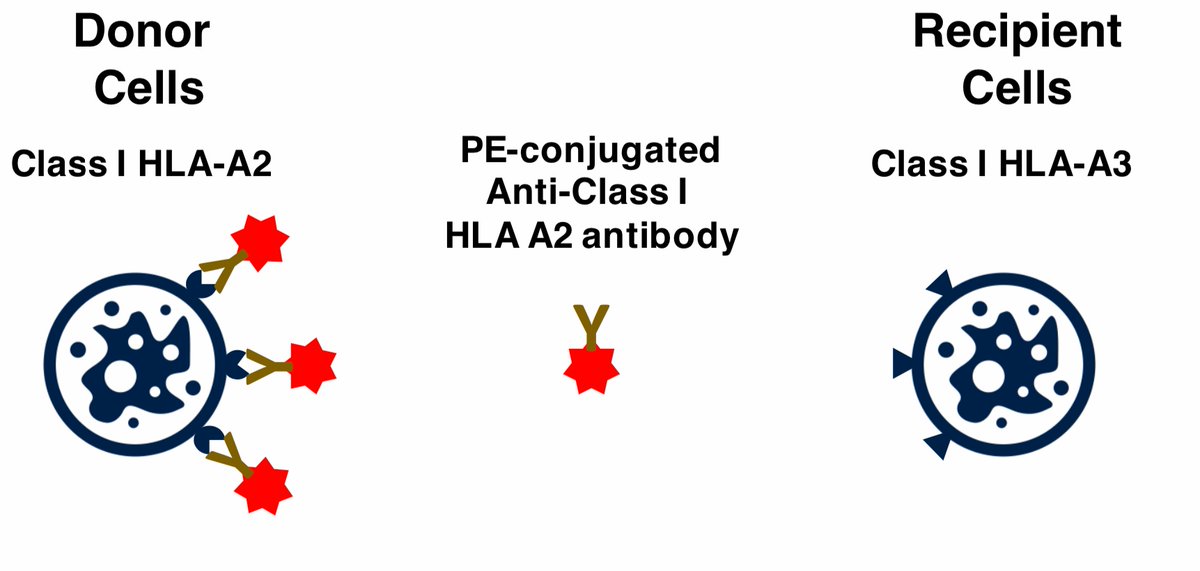
However, a very cool technique - HLA allele congenic cell tracking - allows you to identify definitive Trm cells in transplanted organs. Transplant donor and recipients are often mismatched for Class I HLA alleles, and use antibodies discriminate between them.
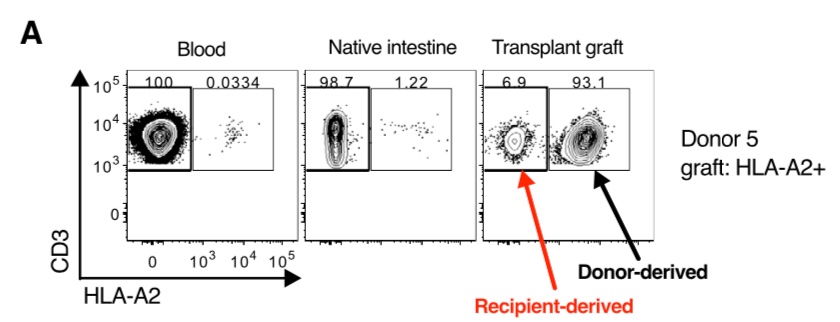
Organ transplantation is now a really exciting context in which to study human Trm cells. Using this technique, we studied human Trm cells in the gut following intestinal transplantation. The technique works really well for some mismatches!
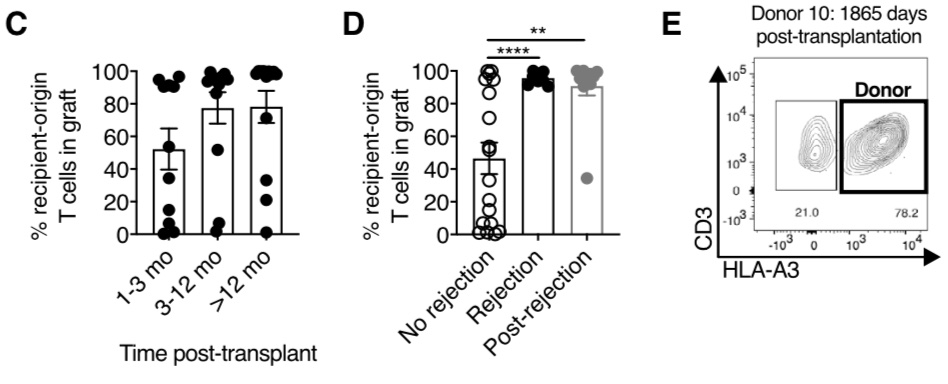
We found donor-derived Trm cells persisting up to 5 years post-transplantation! The persistence is related to episodes of cellular rejection - the donor-derived cells disappear following a rejection episode, although cause and effect is unclear here.
While most Trms were CD103+, but some CD103- populations persisted. We wondered if these were different subsets. What constitutes a true subset is an interesting epistemic issue, but for now let’s say subsets need to be transcriptionally, phenotypically, & functionally distinct.
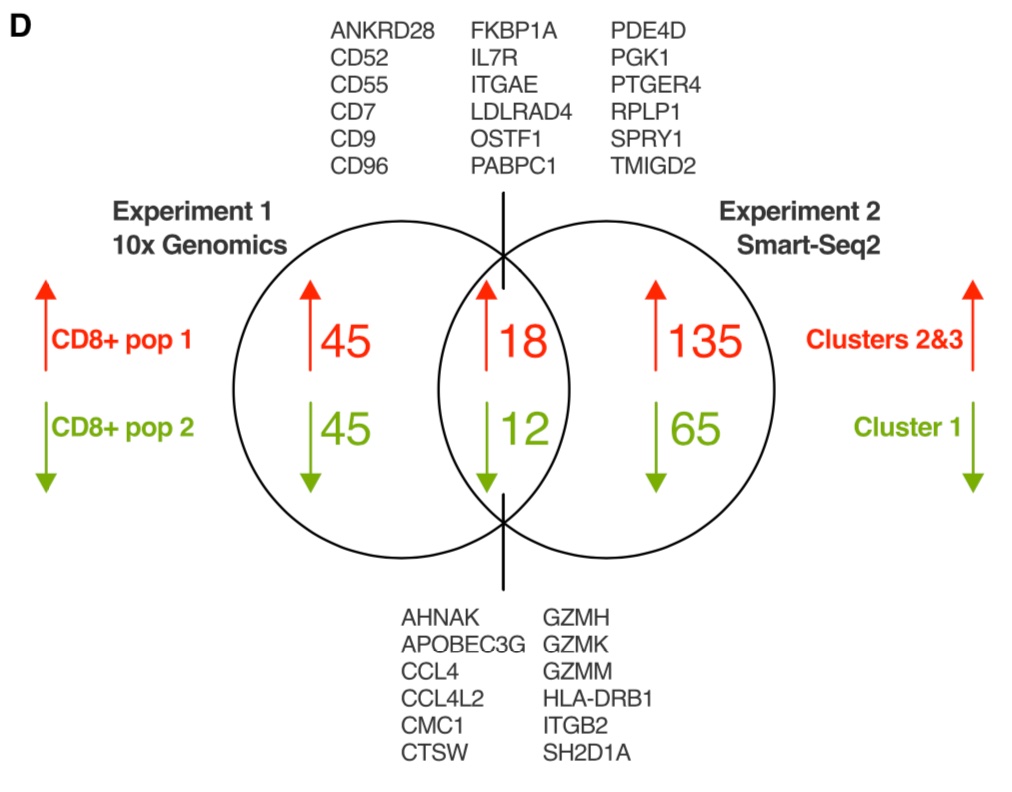
We examined transcriptional heterogeneity using single-cell RNAseq. We identified 2 transcriptionally distinct clusters of CD8+ Trm cells. These differed in expression of integrins, chemokine receptors, and cytotoxic molecules.
One interesting point is that some of these differentially expressed genes have previously been identified in transcriptional signatures of human Trm cells derived from bulk RNAseq, implying that these previous signatures represent an amalgamation of two or more subsets.
We performed a second single-cell RNAseq experiment using a different experimental protocol, which confirmed a core gene set differentially expressed between these two CD8+ Trm cell subsets.
We could identify this gene signature in two other human single-cell RNAseq datasets (including the awesome study from Alison Simmons group below) and even in a recent murine intestinal Trm cell dataset. Therefore this seems to have external validity. https://www.nature.com/articles/s41591-020-1003-4
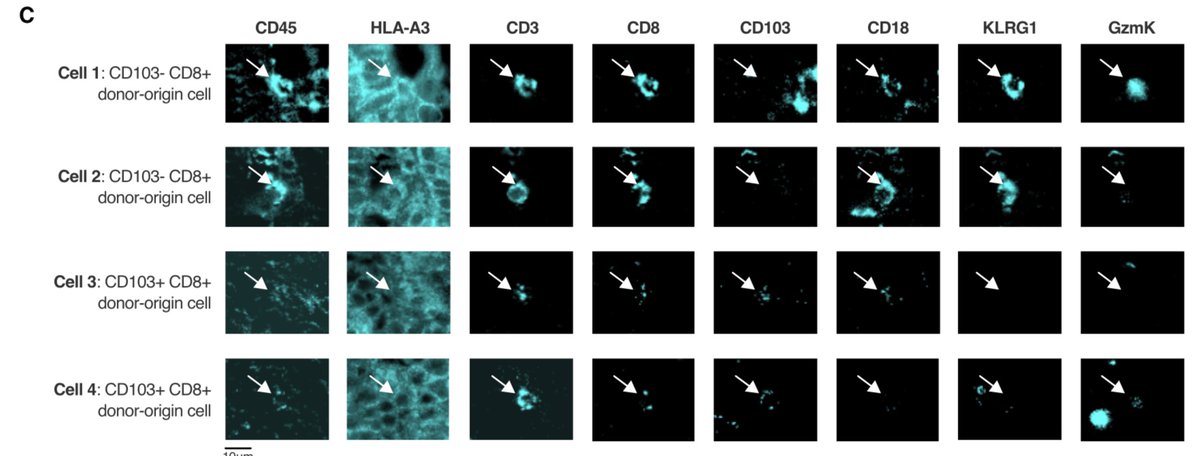
We used flow cytometry, and a cool multiplex imaging technique called chip cytometry, to confirm the presence and phenotypic differences between these two CD8+ Trm cell subsets in both health and intestinal transplantation.
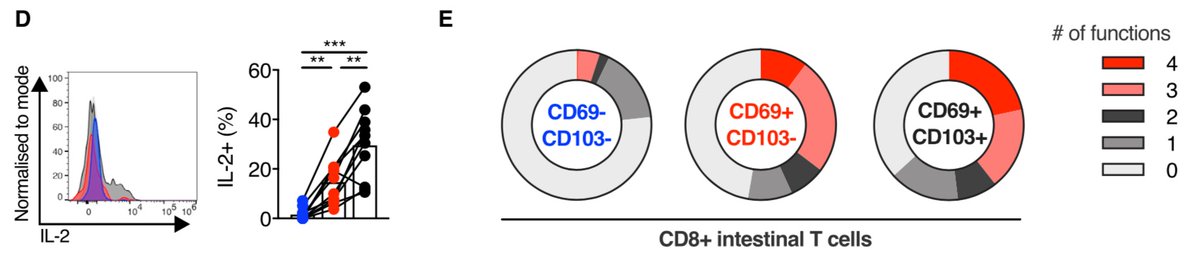
We showed that these are polyfunctional pro-inflammatory populations, with CD103+ CD8+ Trm cells showing increased IL-2 production, which may be related to their long-term persistence similar to hepatic Trm cells (see this paper from @ljpally19) https://rupress.org/jem/article/214/6/1567/42361/IL-2high-tissue-resident-T-cells-in-the-human#.YArYM_MbOIE.twitter
TL/DR: Human CD8+ Trm cells come in (at least) two exciting flavours, which are transcriptionally, phenotypically, & functionally distinct. Lots of potential implications for infective, inflammatory, & neoplastic gut conditions, as well as in the context of organ transplantation.
This has been a super-fun, collaborative, and interesting project. Particular thanks to co-lead authors @TheLucyGarner & @nprovine, as well as Kate Powell, Helen Ferry, @aliamini419 and @sophs012, plus Paul Klenerman, @Ejs17_91 (my awesome supervisors).
Thank you also to the clinicians in the transplant team: Tim, Peter, Georgios and Srikanth, and in particular the fantastic Phil Allan, gastroenterologist at the Oxford intestinal transplant programme, for the leadership and work to bring this collaborative project off.
Finally I thank the patients who consented for this research. Intestinal transplantation is tough, and they have been through so much. To volunteer for this is such a gift, a way to pay forward to future patients who will benefit from their contribution. Absolute legends.

 Read on Twitter
Read on Twitter