A  on T cell receptor repertoire diversity in cancer RNA-sequencing studies, based on great work by @BortoneDante reported in @CIR_AACR: …https://cancerimmunolres-aacrjournals-org.libproxy.lib.unc.edu/content/9/1/103 (1/n)
on T cell receptor repertoire diversity in cancer RNA-sequencing studies, based on great work by @BortoneDante reported in @CIR_AACR: …https://cancerimmunolres-aacrjournals-org.libproxy.lib.unc.edu/content/9/1/103 (1/n)
 on T cell receptor repertoire diversity in cancer RNA-sequencing studies, based on great work by @BortoneDante reported in @CIR_AACR: …https://cancerimmunolres-aacrjournals-org.libproxy.lib.unc.edu/content/9/1/103 (1/n)
on T cell receptor repertoire diversity in cancer RNA-sequencing studies, based on great work by @BortoneDante reported in @CIR_AACR: …https://cancerimmunolres-aacrjournals-org.libproxy.lib.unc.edu/content/9/1/103 (1/n)
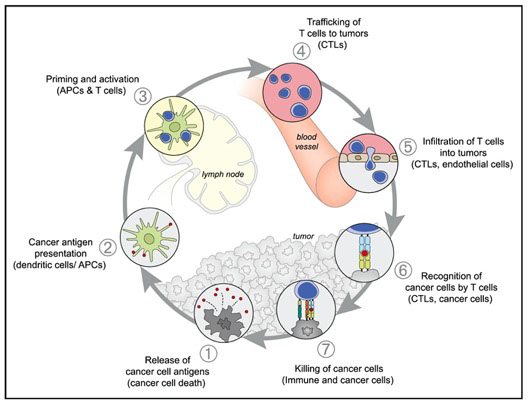
T cells killing tumor cells is a final common pathway in immuno-oncology. Many immunotherapies, including immune checkpoint inhibitors and therapeutic neoantigen vaccines, depend on the endogenous T cell population to be stimulated then eliminate cancer cells. (2/n)
T cell receptor repertoire diversity has been shown to be both positively and negatively correlated with response to immunotherapy, highlighting the complexity of TCR repertoire biology. TCR repertoire diversity is also notoriously difficult to measure. (4/n)
Multiple mathematical indices of diversity are reported, and repertoire profiling experiments suffer from multi-level undersampling: tissue samples don't represent the whole organism repertoire or the whole of tissue heterogeneity... (5/n)
...and repertoire profiling methods often fail to cover the whole repertoire in the tissue sample itself. We sought to determine the performance of diversity indices in the context of undersampling, so we analyzed data simulated using STIG ( https://www.biorxiv.org/content/10.1101/2020.02.28.969469v1) (6/n)
All diversity indices were effected by undersampling, with decreased error as the sampling improved. (7/n)
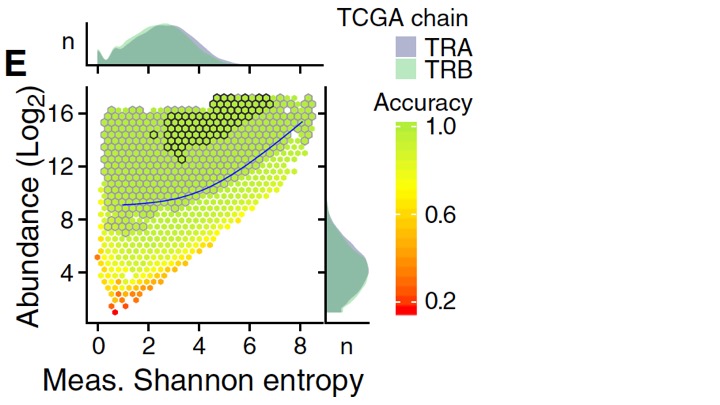
To determine conditions under which we could trust the TCR repertoire measurements, we evaluated diversity estimation error by abundance (number of TCR reads in the RNA-seq data) and shannon entropy (our favorite index of diversity). (8/n)
Under conditions of high abundance and low entropy, the direct calculation of the shannon entropy value was quite accurate. Unfortunately, RNA sequencing experiments often have TCR read counts low enough to make diversity calculations unreliable. (9/n)
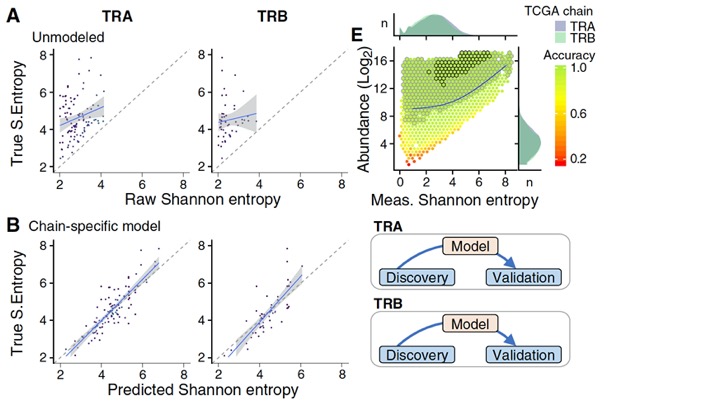
We wanted to correct the errors in diversity estimation to derive a more reproducible diversity calculation for use in biomarker development. To do this, we split our simulated data into a 2/3 discovery set and 1/3 validation set, ... (10/n)
...applied elastic net linear regression with monte carlo cross-validation to build a model from diversity features trained on the discovery set, then assessed model performance on the validation set. Diversity estimation improved. (11/n)
The modeling approach gave improved performance even when trained in an alpha/beta chain agnostic fashion. (12/n)
There was discrimination of survival and response differences in the Riaz et al. melanoma cohort ( https://www.sciencedirect.com/science/article/pii/S0092867417311224?via%3Dihub) when our estimated diversity was used compared to direct diversity calculation. (13/n)
We are working on prospective validation of this diversity estimation approach, and we hope it will be useful to researchers doing TCR repertoire profiling from tumor RNA-seq data. (14/n)
We appreciate funding support from @UNC_Lineberger, @SusanGKomen, @TheVFoundation, and @CancerResearch for supporting diversity estimation on the @iatlas_cri platform. Also thank you to the reviewers and excellent editorial support at @CIR_AACR, and thank you for reading! (fin)

 Read on Twitter
Read on Twitter