Whether homologous recombination has a net beneficial or detrimental effect on adaptive evolution is largely unexplored in natural bacterial populations. https://www.biorxiv.org/content/10.1101/2021.01.20.427438v1
We address this question by evaluating polymorphism and divergence data across 196 bacterial genome sequences of five closely-related Rhizobium leguminosarum species (genomes previously assembled and published here: https://twitter.com/izabelcavassim/status/1240954105193005057)
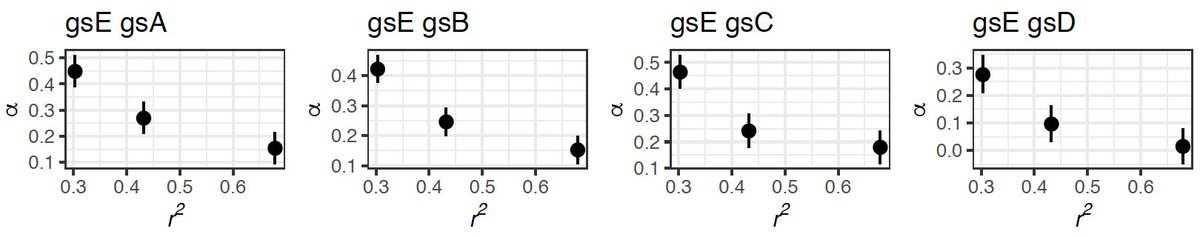
We show that the proportion of amino acid changes fixed due to adaptive evolution (𝛼) increases with an increased recombination rate. This relationship is observed in the interspecific levels––in which the most recombining species has the highest 𝛼.
We then evaluate the impact of recombination within each species by dividing genes into three equally sized recombination classes based on their average level of intragenic linkage disequilibrium (r^2). And observed a similar pattern: higher 𝛼 in the more recombining class
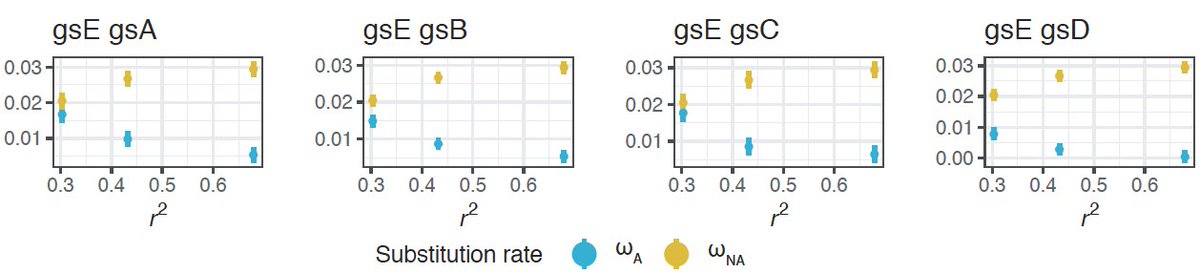
This is both due to a higher estimated rate of adaptive evolution (⍵𝑎) and a lower estimated rate of non-adaptive evolution (⍵𝑛𝑎), suggesting that recombination both increases the fixation probability of advantageous variants and decreases the fix. prob. of deleterious ones.
These results suggest that recombination facilitates adaptive evolution in prokaryotes, as it has been shown previously to be in eukaryotes. Thanks to @mikkelschierup @ThomasBataillon and @stiguandersen for this collaboration and https://genome.au.dk/ for the computing facility

 Read on Twitter
Read on Twitter