A little late to the party (blame it on time zones), but I'm happy to share our initial findings on the reactivation of H2A.B in cancers, from an international collaboration with @AntoineMolaro and @JSarthy . #3amigos. https://www.nature.com/articles/s41467-020-20707-x
Many thanks to our current/former/forever mentors @bradleybio, @HarmitMalik and Steve Henikoff for supporting this endeavour that took off at the end of our post-doc stints at the @fredhutch.
This project emerged from work to characterize the molecular roles of cancer-specific genes: genes that are expressed in cancers, but not in normal tissue.
DUX4, a developmental transcription factor, was the first of these genes that we characterized. https://www.cell.com/developmental-cell/fulltext/S1534-5807(19)30529-5
DUX4, a developmental transcription factor, was the first of these genes that we characterized. https://www.cell.com/developmental-cell/fulltext/S1534-5807(19)30529-5
DUX4 is one of many such genes. Why did we turn our attention H2A.B next?
Because of a poster session at the @Basic Sciences retreat (trainees take note).
I was browsing the @HarmitMalik lab posters, and noticed a familiar gene ID on @AntoineMolaro 's poster.
Because of a poster session at the @Basic Sciences retreat (trainees take note).
I was browsing the @HarmitMalik lab posters, and noticed a familiar gene ID on @AntoineMolaro 's poster.
@AntoineMolaro had been investigating the in vivo endogenous functions H2A.B, a multi-copy histone variant bearing interesting evolutionary signatures.
Some cool results of that work published here: https://genome.cshlp.org/content/28/4/460 and https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001001
Some cool results of that work published here: https://genome.cshlp.org/content/28/4/460 and https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001001
More recently, @JSarthy brought #oncohistones to our attention.
Recent work by the Allis and @jdlicht groups highlighted recurrent mutations in cancer within canonical histones that affect nucleosome stability.
https://www.nature.com/articles/s41586-019-1038-1 and https://cancerdiscovery.aacrjournals.org/content/9/10/1438
Recent work by the Allis and @jdlicht groups highlighted recurrent mutations in cancer within canonical histones that affect nucleosome stability.
https://www.nature.com/articles/s41586-019-1038-1 and https://cancerdiscovery.aacrjournals.org/content/9/10/1438
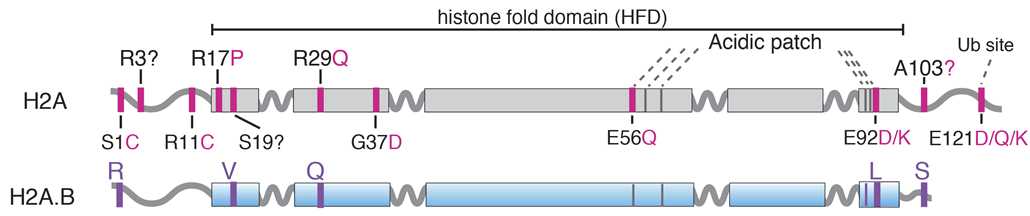
Turns out that H2A.B looks a lot like the mutated canonical H2A, suggesting that reactivated H2A.B and mutated H2A may converge on similar molecular mechanisms to promote cancer.
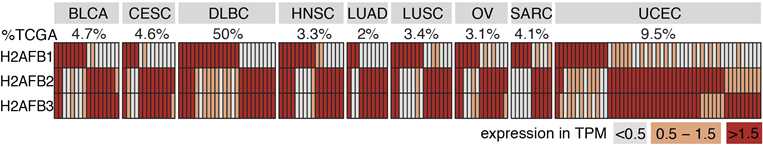
We found that H2A.B is reactivated in many cancer types, notably in ~half of DLBCLs, and likely other lymphomas as well. This was also recently reported by others: https://www.biorxiv.org/content/10.1101/2021.01.19.427265v1
In fact, individual tumors reactivated different paralogous copies of H2A.B (put a pin in that).
Hold up... what is H2A.B again?
H2A.B is a truncated version of the canonical H2A that exists in a few copies in the genome (on the X chromosome).
Incorporation of H2A.B in nucleosomes has been shown to destabilize them.
H2A.B is a truncated version of the canonical H2A that exists in a few copies in the genome (on the X chromosome).
Incorporation of H2A.B in nucleosomes has been shown to destabilize them.
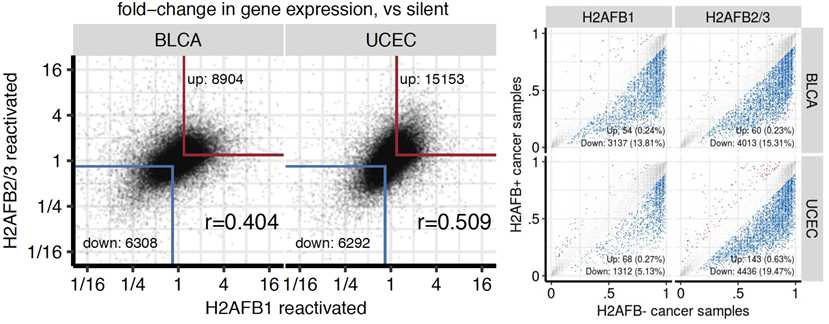
We find that H2A.B expression is associated with widespread gene dysregulation.
This gene dysregulation is context-specific: different cancer types have different up/down-regulated genes with H2A.B expression (even if they are consistent within a cancer type).
This gene dysregulation is context-specific: different cancer types have different up/down-regulated genes with H2A.B expression (even if they are consistent within a cancer type).
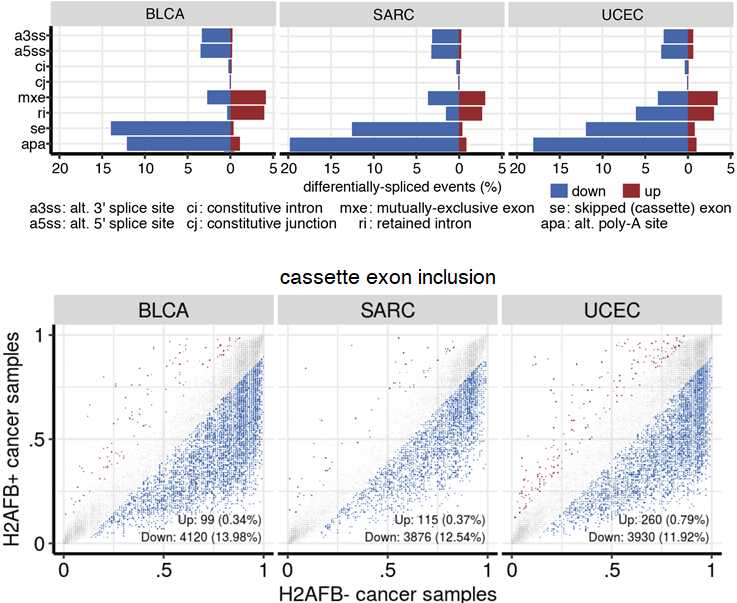
Strikingly, we observe widespread and consistent alternative splicing dysregulation in H2A.B expressing cancers.
In fact, the extent of splicing dysregulation exceeds what we've previously seen in cancers with splicing factor mutations.
In fact, the extent of splicing dysregulation exceeds what we've previously seen in cancers with splicing factor mutations.
My favourite bit of this paper: how could we show that H2A.B likely *caused* this gene dysregulation by computational methods?
Remember the various paralogs of H2A.B being reactivated in different individual tumors?
Remember the various paralogs of H2A.B being reactivated in different individual tumors?
We found the same gene expression and alternative splicing dysregulation in cancers that independently reactivated different H2A.B paralogs.
Moreover, the patterns of gene dysregulation are consistent with unstable nucleosomes affecting splicing (co-transcriptionally).
Moreover, the patterns of gene dysregulation are consistent with unstable nucleosomes affecting splicing (co-transcriptionally).
Much remains to be explored, including why so many lymphomas reactivate H2A.B, how H2A.B promotes cancer, and whether we can take advantage of H2A.B-driven gene/splicing dysregulation to treat such cancer patients.
This has been an immensely fun and enjoyable collaboration with @JSarthy and @AntoineMolaro, conducted remotely over 3 time zones, in the midst of COVID-19 lockdowns in Seattle, France and Singapore. Looking forward to more exciting discoveries together!

 Read on Twitter
Read on Twitter