There's been a lot of news today about 'new variants in Brazil' & 'S:E484' and 'Ohio variants'. A short thread discussing what we know about each.
1/11
#SARSCoV2 #Ohio #BrazilVariant #Brazil #COVID19
1/11
#SARSCoV2 #Ohio #BrazilVariant #Brazil #COVID19
There are two variants circulating in Brazil: 20J/501Y.V3, recently detected in Japan & prevalent in Manaus. And temporarily-labelled '20B/S.484K', a larger clade circulating more widely in Brazil.
They both carry the spike mutation 484K, though this arise separately.
2/11
They both carry the spike mutation 484K, though this arise separately.
2/11
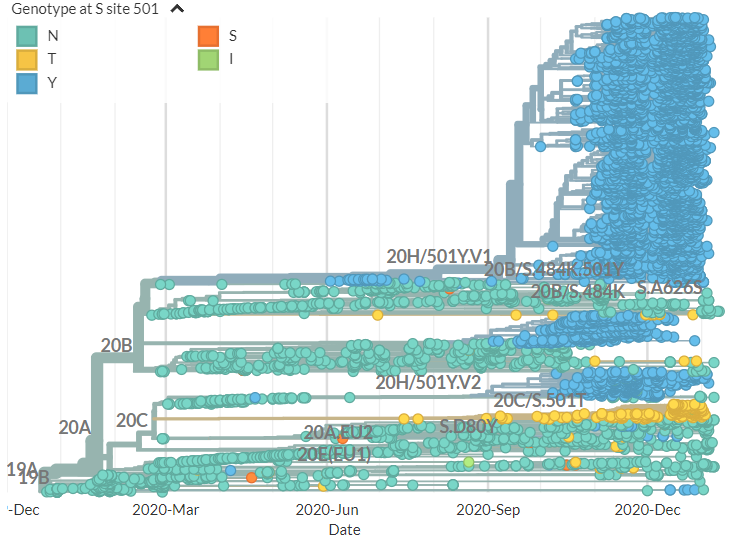
S:E484K is also found in 501Y.V2 (primarily circulating in South Africa).
Why are we particularly concerned about S:E484K? You can read more at http://CoVariants.org , as discussed in a tweet from earlier this week:
3/11 https://twitter.com/firefoxx66/status/1348939611578503168
Why are we particularly concerned about S:E484K? You can read more at http://CoVariants.org , as discussed in a tweet from earlier this week:
3/11 https://twitter.com/firefoxx66/status/1348939611578503168
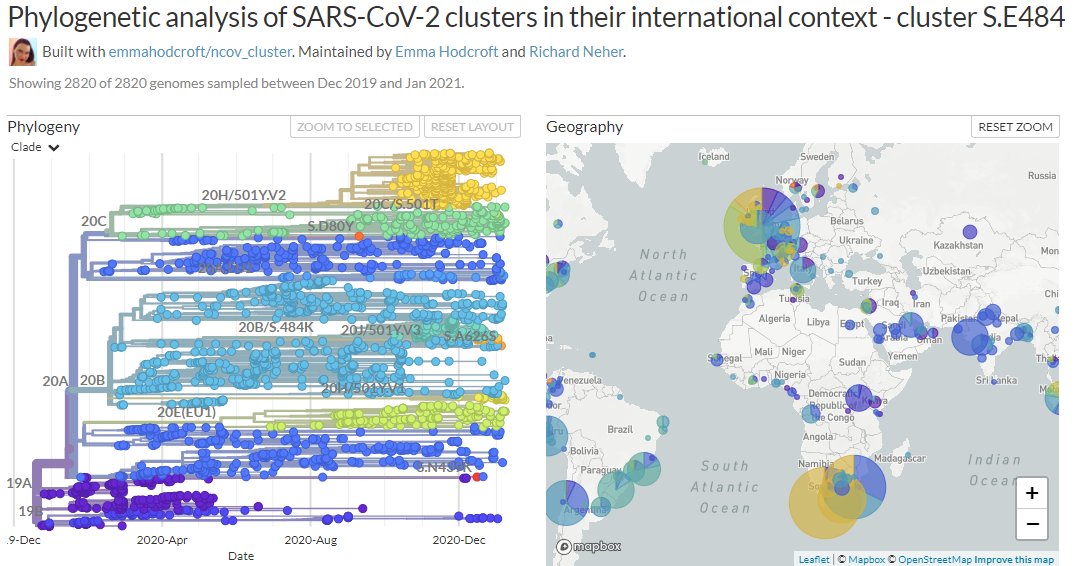
The focal build for S:E484 is updated with data from 13 Jan & includes the sequences from Manaus, Brazil which cluster with the recent Japanese travel sequences in new Nextstrain clade 20J/501Y.V3:
https://nextstrain.org/groups/neherlab/ncov/S.E484
Read about the Manaus seqs:
https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586
4/11
https://nextstrain.org/groups/neherlab/ncov/S.E484
Read about the Manaus seqs:
https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586
4/11
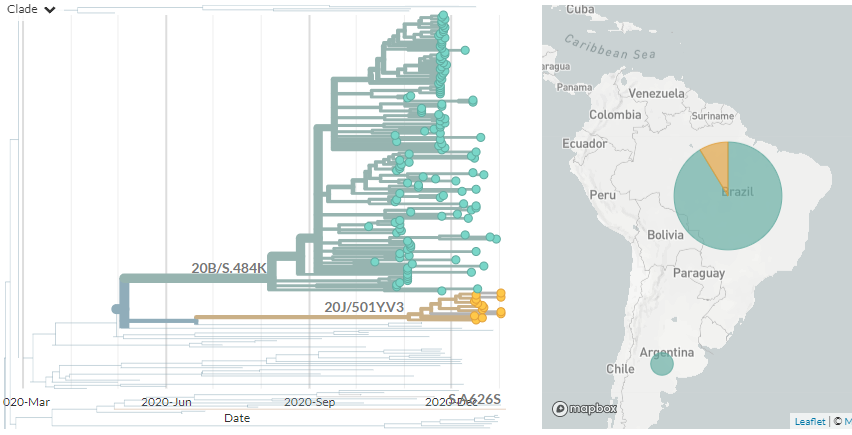
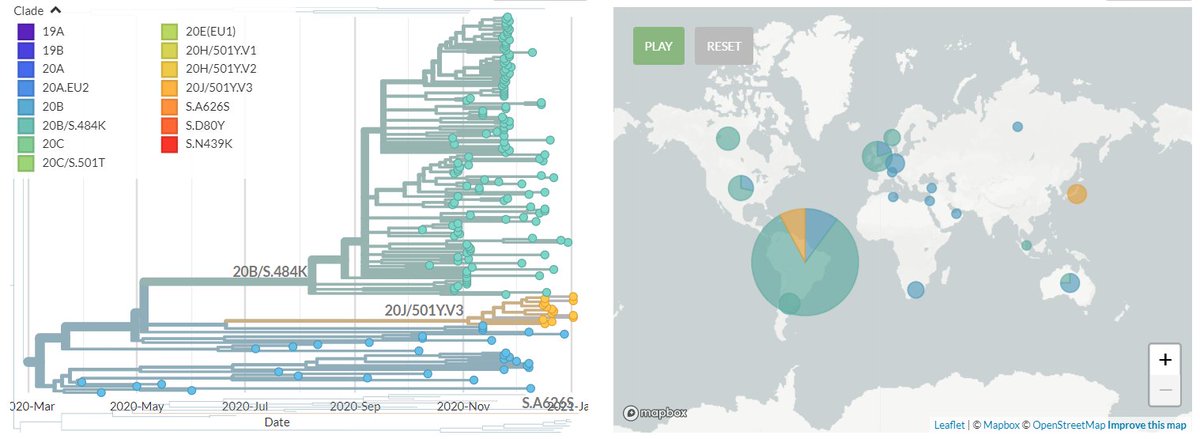
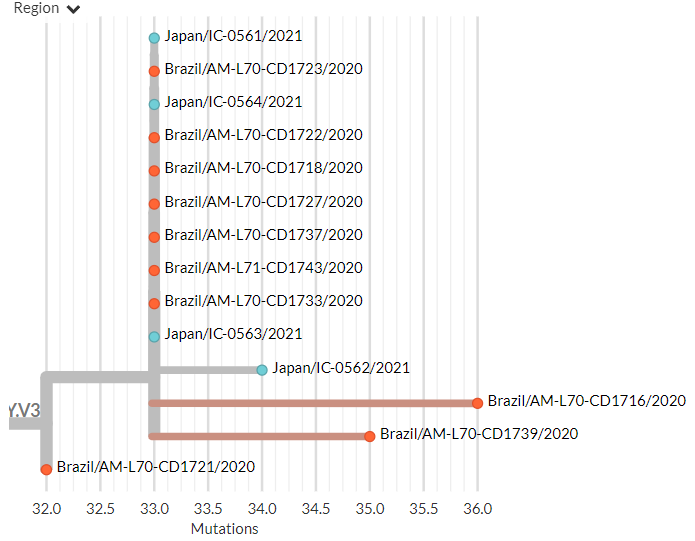
20J/501Y.V3 sequences have both 484K & 501Y (like 501Y.V2, mostly in South Africa) - they are shown in orange below.
They are clearly separate from the other, larger S:E484K cluster circulating in Brazil (in green, with temporary label 20B/S.484K)
5/11
They are clearly separate from the other, larger S:E484K cluster circulating in Brazil (in green, with temporary label 20B/S.484K)
5/11
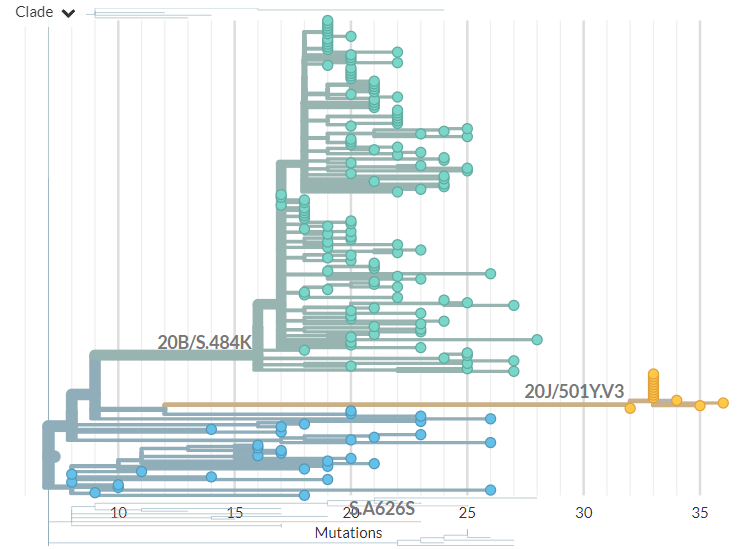
In divergence (mutation) view, the 501Y.V3 variant has a 'long branch' - it sticks out to the right. This means it has a high number of mutations relative to surrounding sequences.
Zooming in, we can see that the Japanese samples are nested within the Brazilian diversity.
6/11
Zooming in, we can see that the Japanese samples are nested within the Brazilian diversity.
6/11
There have also been some news reports circulating about 2 'Ohio variants'.
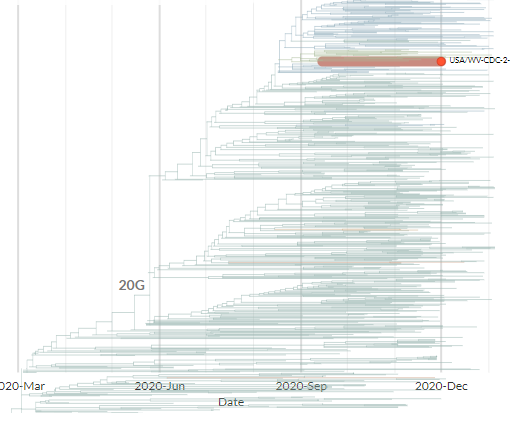
From press release data, one is a 20G clade sequence with S:N501Y. As shown below (blue) 501Y mutations have popped up in the past. While worth keeping an eye on, I think this isn't so remarkable.
7/11
From press release data, one is a 20G clade sequence with S:N501Y. As shown below (blue) 501Y mutations have popped up in the past. While worth keeping an eye on, I think this isn't so remarkable.
7/11
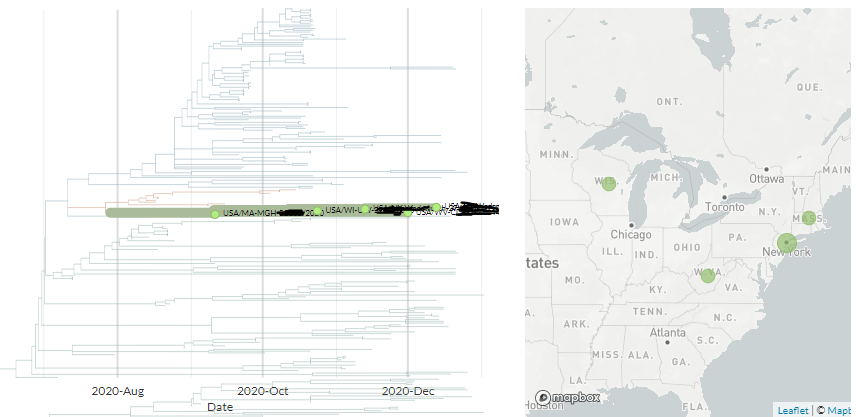
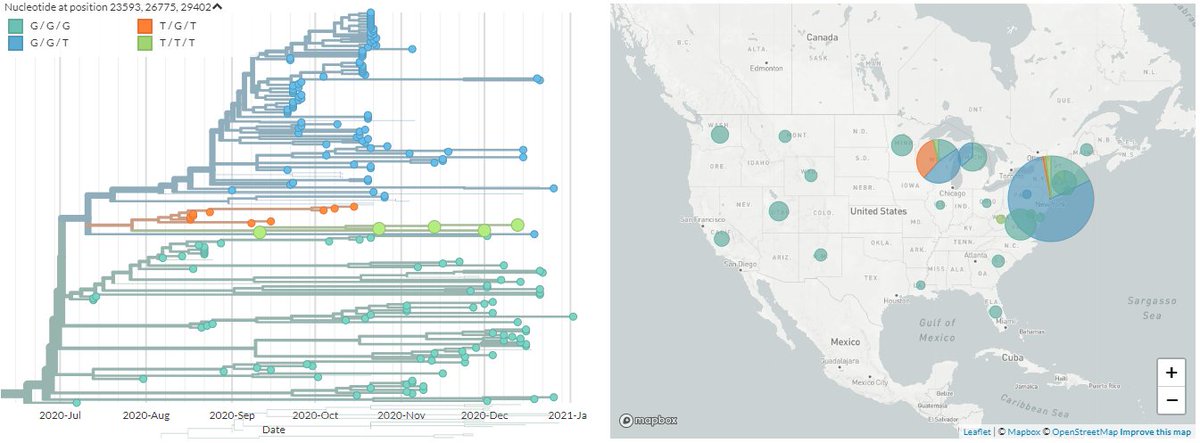
The other variant identified seems to have 3 mutations, 1 each in the Spike, Membrane, & Nucleocapsid proteins in a 20G background. There are 17 USA seqs with the same mutations (Only 1 of these is currently visible in N America Nextstrain build).
8/11
8/11
The Ohio samples are not yet online, but the 17 existing samples that seem to have the same mutations are from Massachusetts, Maryland, Minnesota, New York, Texas, & Wisconsin.
Another USA-state focused build captures 5 of them, all dating between 9 Dec & 12 Jan.
9/11
Another USA-state focused build captures 5 of them, all dating between 9 Dec & 12 Jan.
9/11
The preprint states that in Columbus, OH, at end of Dec, the variant went from 10% prevalence to 60% prevalence (10-20 samples per week) in 3 weeks.
I'd be curious to hear if other states have seen this variant & if so, also if they've seen similar rises.
10/11
I'd be curious to hear if other states have seen this variant & if so, also if they've seen similar rises.
10/11
I have been careful here not to mention the mutations as, so far as I can tell, the preprint is not yet out. I'll update with a link to the preprint & the exact mutations when I hear it's up.
You can read more below:
https://news.osu.edu/new-variant-of-covid-19-virus-discovered-in-columbus/
https://www.nytimes.com/live/2021/01/13/world/covid19-coronavirus/researchers-in-ohio-find-preliminary-evidence-of-two-virus-variants-that-may-be-more-contagious
11/11
You can read more below:
https://news.osu.edu/new-variant-of-covid-19-virus-discovered-in-columbus/
https://www.nytimes.com/live/2021/01/13/world/covid19-coronavirus/researchers-in-ohio-find-preliminary-evidence-of-two-virus-variants-that-may-be-more-contagious
11/11
The preprint is still not out, but the mutations seem to be common knowledge on Twitter - so they are S:Q677H, M:A85S, N:D377Y, or nucleotides 23593, 26775, 29402.
These are present in around 17 sequences in the US, though not all show up in the same Nextstrain builds.
12/11
These are present in around 17 sequences in the US, though not all show up in the same Nextstrain builds.
12/11
You can see the cluster I picture up-thread in the SPHERES New York build - mouse-over 'T/T/T' in legend to see them.
13/11
https://nextstrain.org/groups/spheres/ncov/new-york?c=gt-nuc_23593,26775,29402&f_country=USA
13/11
https://nextstrain.org/groups/spheres/ncov/new-york?c=gt-nuc_23593,26775,29402&f_country=USA

 Read on Twitter
Read on Twitter