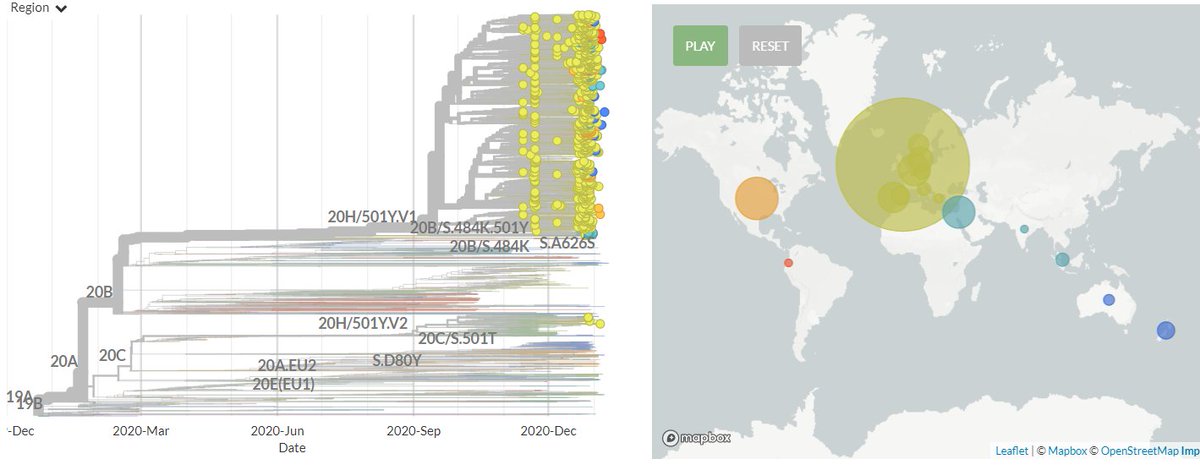
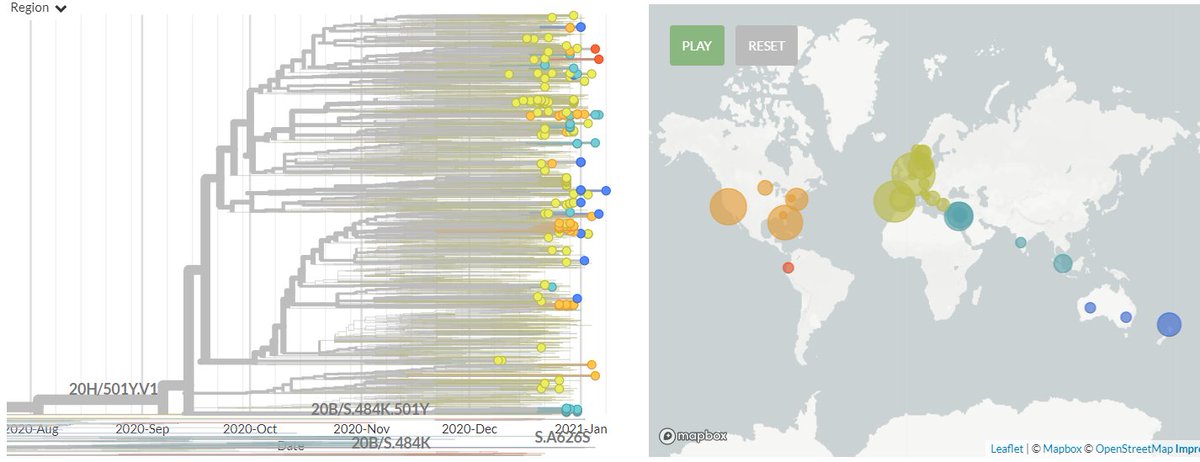
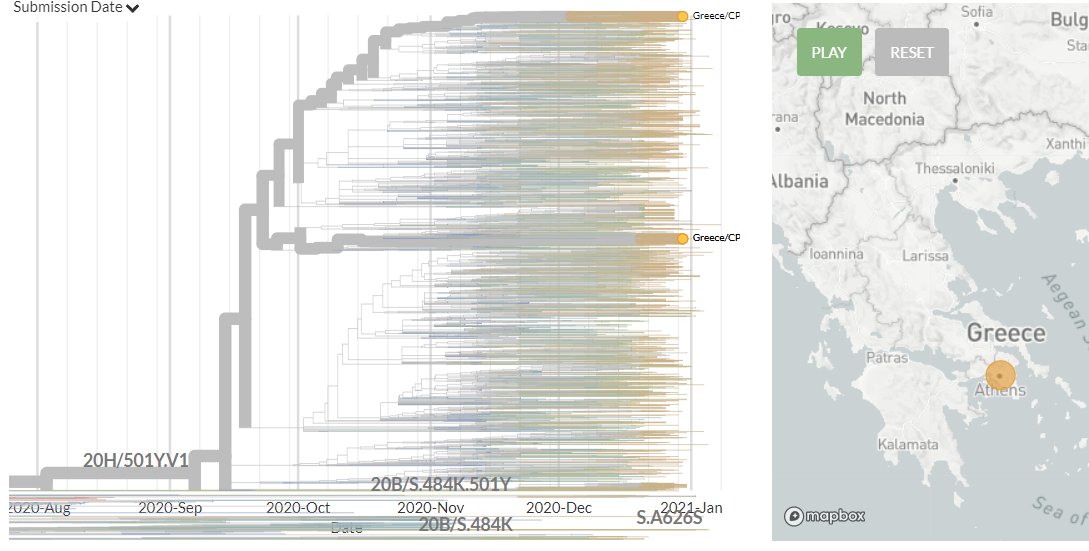
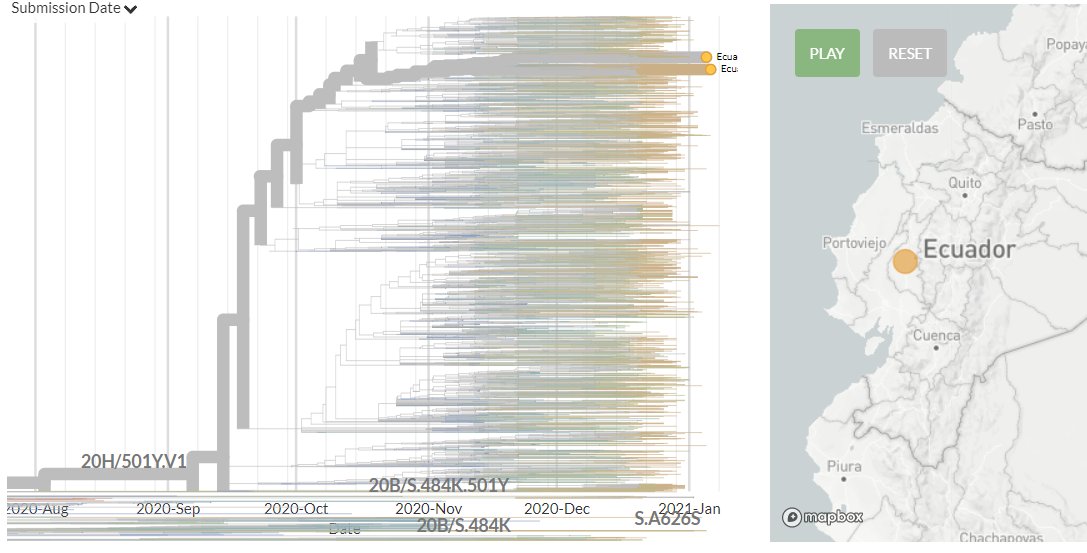
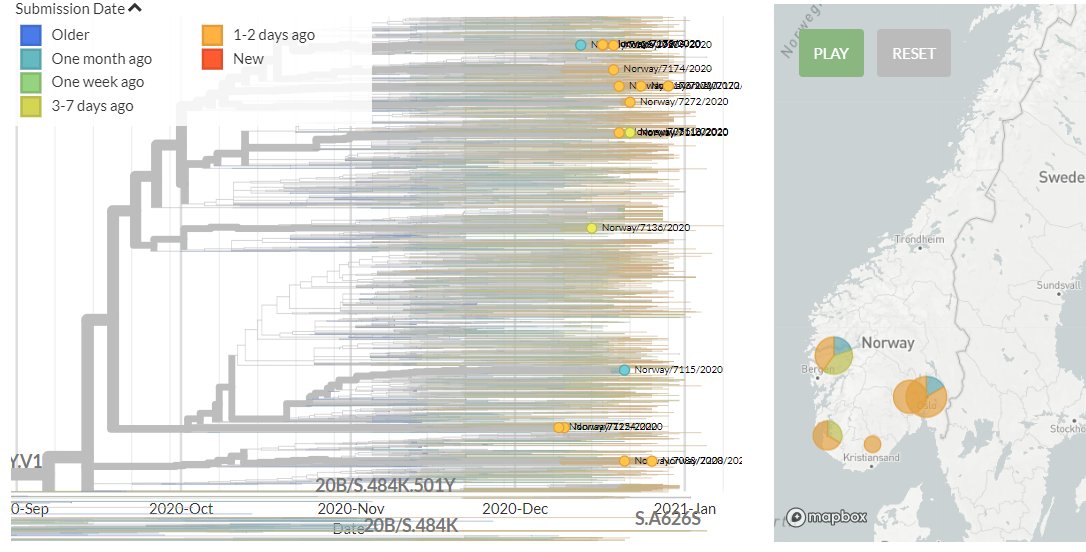
The focal S:N501 build is now updated with data from 13 Jan!
There are a total of 247 non-UK & non-South African sequences in 501Y.V1 (B.1.1.7 #b117) & 501Y.V2.
I'll do an additional thread later on, covering S:E484 & the 'Ohio variants'.
1/18
https://nextstrain.org/groups/neherlab/ncov/S.N501?p=grid
There are a total of 247 non-UK & non-South African sequences in 501Y.V1 (B.1.1.7 #b117) & 501Y.V2.
I'll do an additional thread later on, covering S:E484 & the 'Ohio variants'.
1/18
https://nextstrain.org/groups/neherlab/ncov/S.N501?p=grid
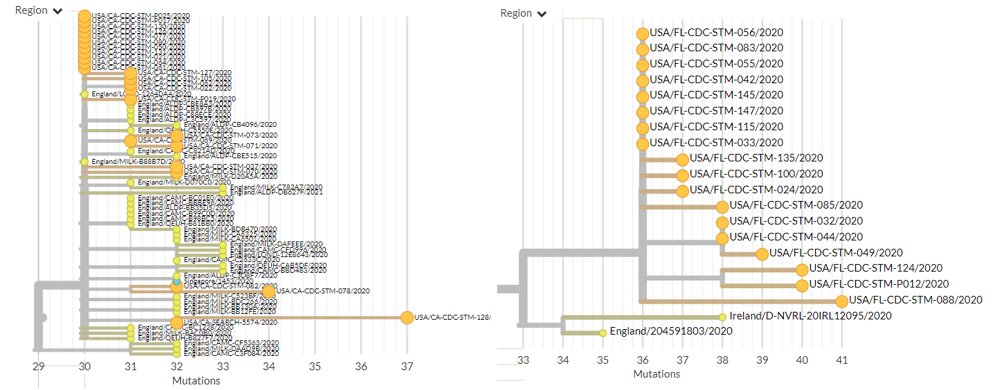
There are 244 new sequences in 501Y.V1 (B.1.1.7) from Australia, Belgium, Denmark, Germany, India, Israel, Italy, Netherlands, New Zealand, Norway, Portugal, Singapore, Spain & USA, as well sequences as from Ecuador and Greece for the first time.
2/18
2/18
Greece has 3 new sequences in 501Y.V1 for the first time. They indicate 2 separate introductions.
Ecuador has 2 sequences for the first time. They also indicate 2 separate introductions.
3/18
Ecuador has 2 sequences for the first time. They also indicate 2 separate introductions.
3/18
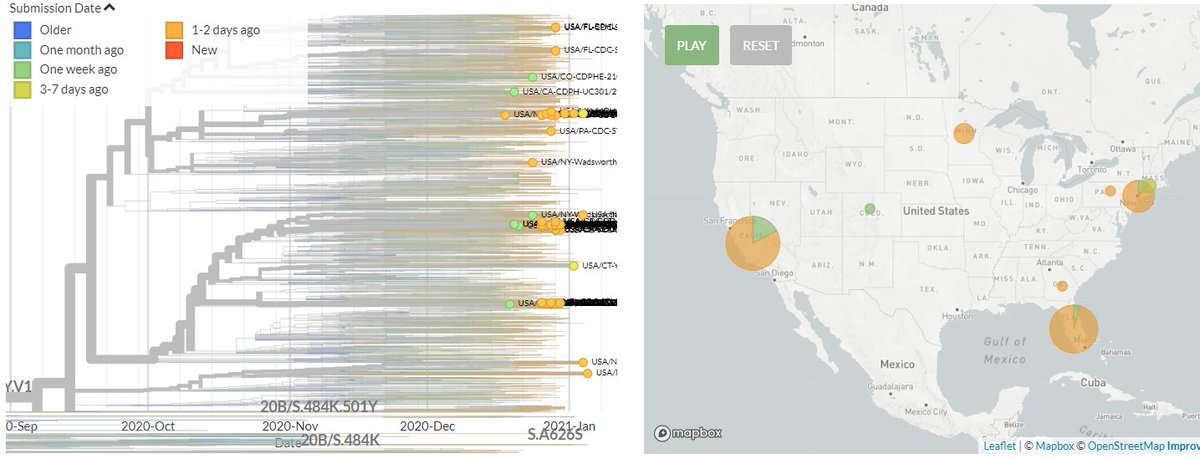
The USA has 61 new sequences (orange), from Florida, Minnesota, California, Pennsylvania, & New York.
A few of these represent separate introductions. Others form distinct clusters that may indicate ongoing local transmission.
4/18
A few of these represent separate introductions. Others form distinct clusters that may indicate ongoing local transmission.
4/18
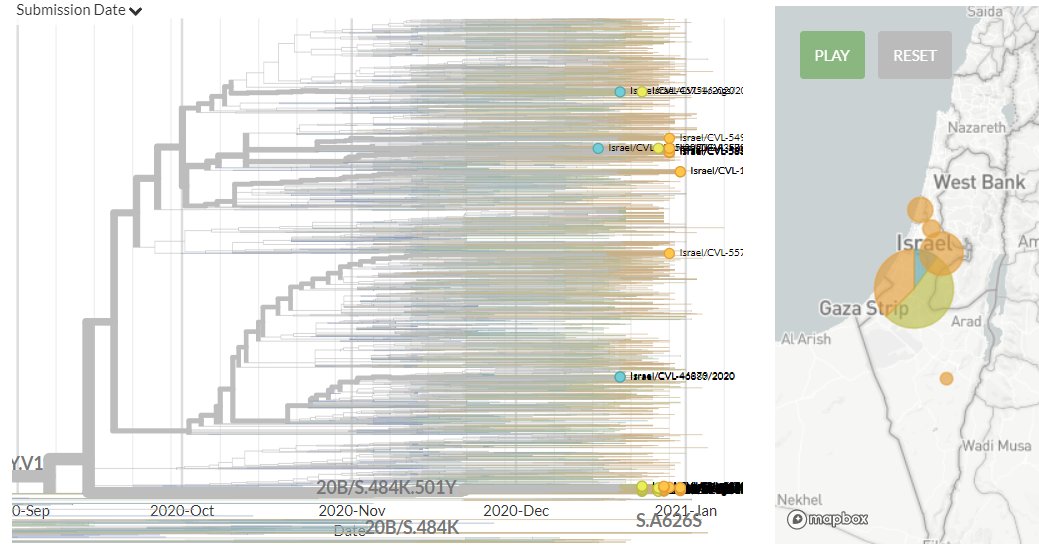
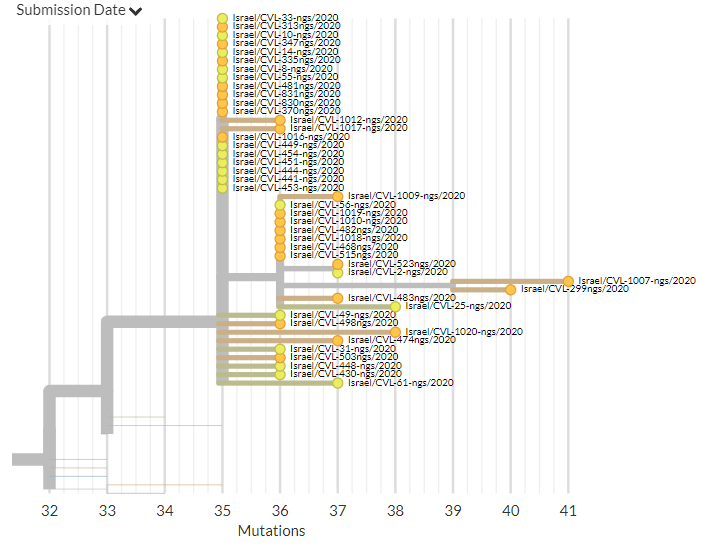
Israel has 35 new sequences (orange). Some of these show separate introductions, but many link with older sequences, indicating ongoing local transmission.
5/18
5/18
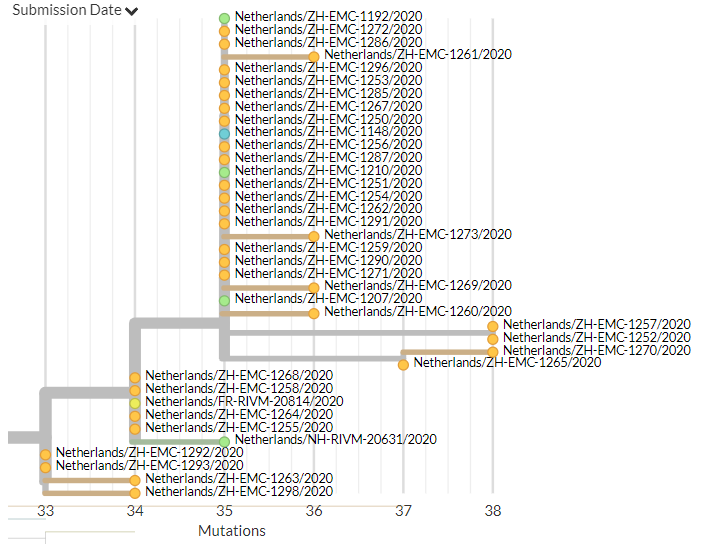
The Netherlands has 34 new sequences (orange). A few of these represent separate introductions, but most form a large cluster with older sequences from the Netherlands, indicating ongoing local transmission.
6/18
6/18
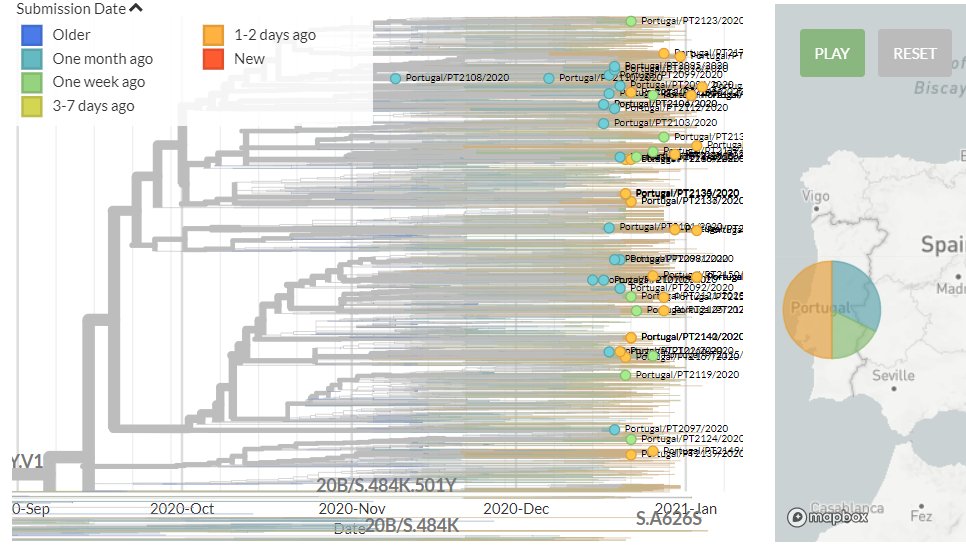
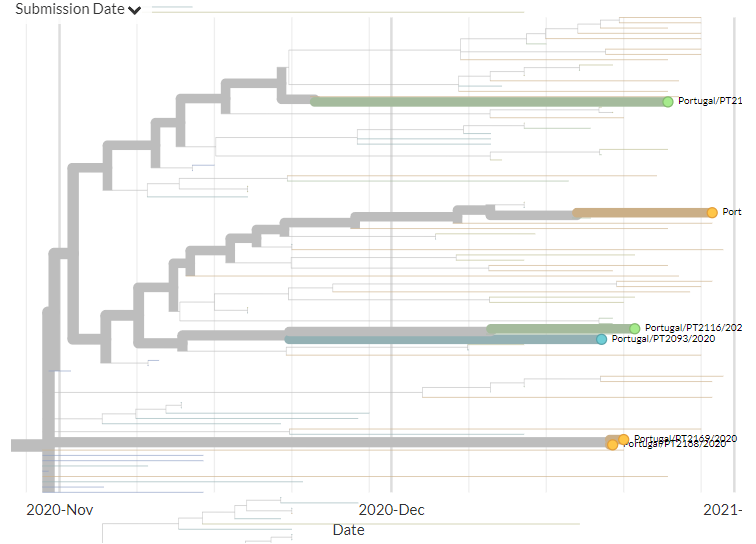
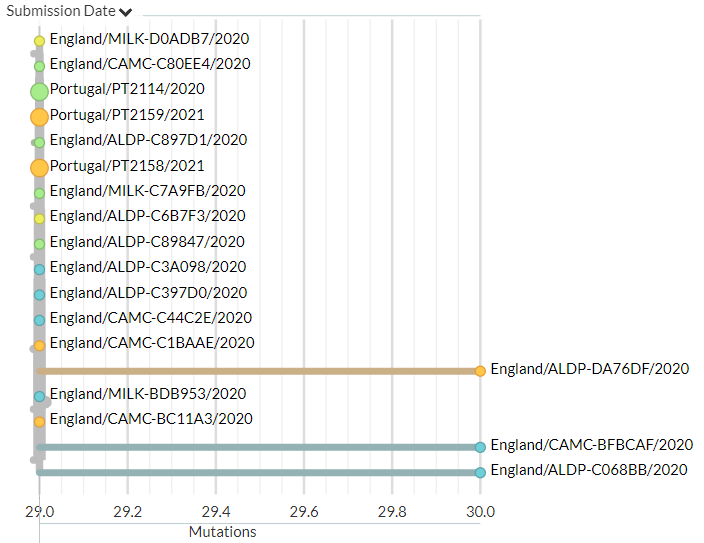
Portugal has 31 new sequences (orange). Though hard to see in the zoomed-out view, most of these represent separate introductions. There is one case where 2 new seqs are identical to an older one, but this could be a common exposure source.
7/18
7/18
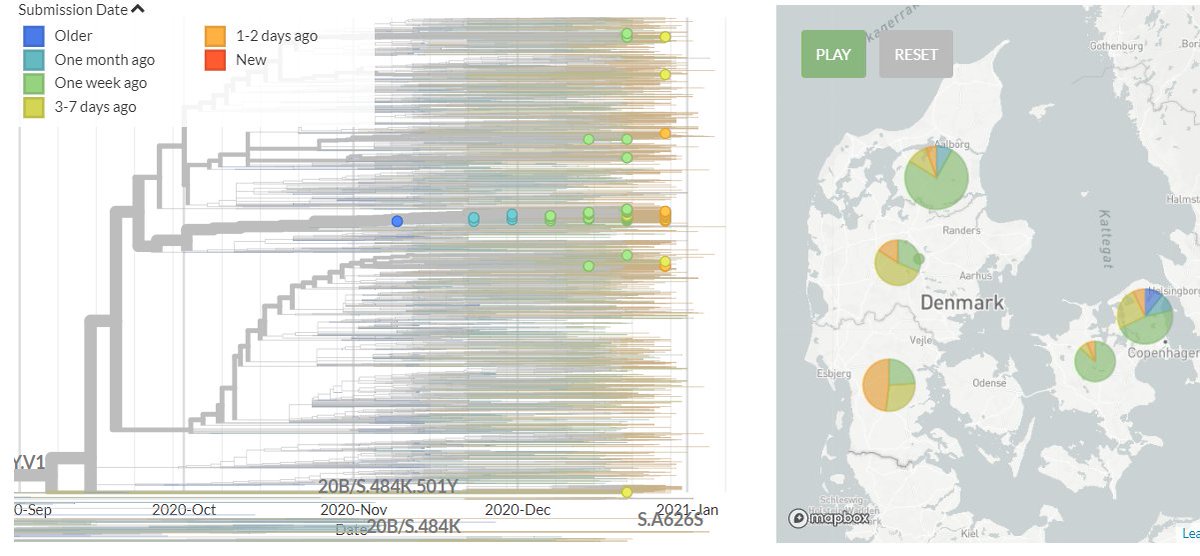
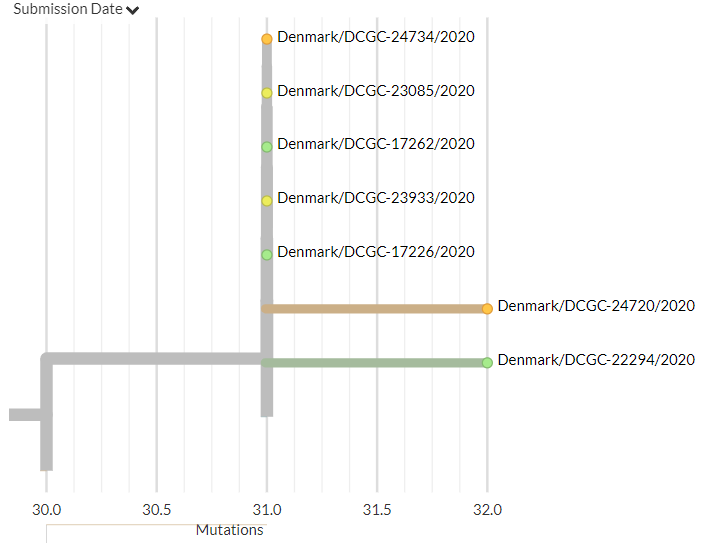
Denmark has 20 new sequences (orange). A few of these indicate separate introductions, but must cluster with older sequences in smaller groups or in the very large Danish group previously identified, indicating local transmission.
8/18
8/18
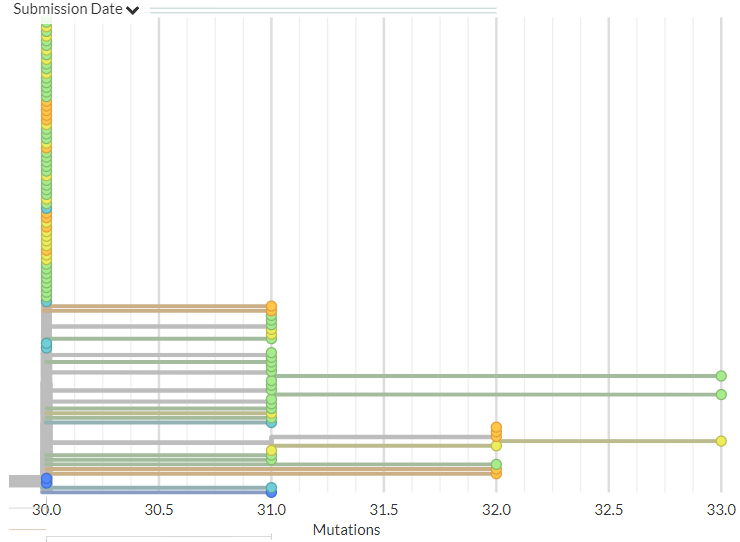
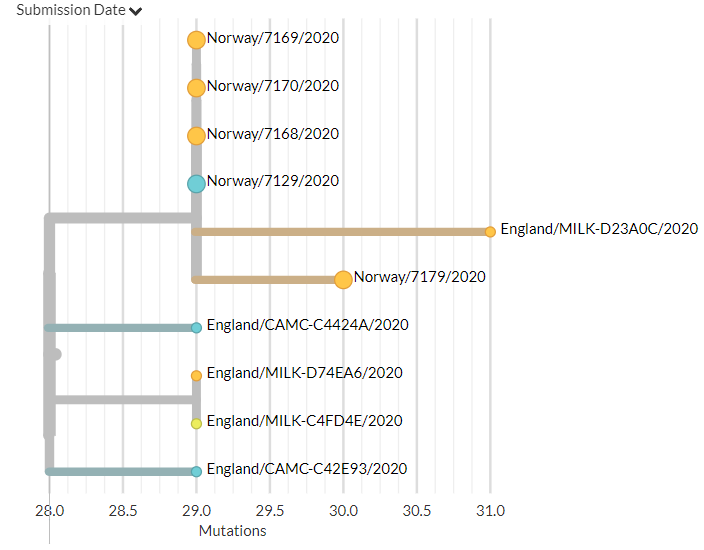
Norway has 14 new sequences (orange). Most indicate separate introductions, but a few are identical to or related to an older sequence, which may indicate local transmission.
9/18
9/18
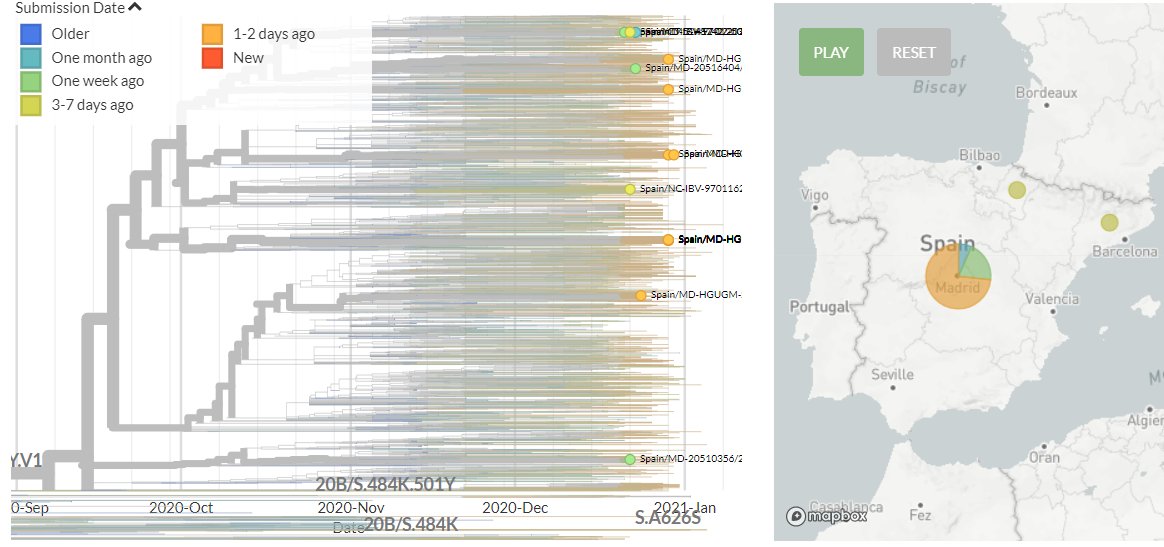
Spain has 11 new sequences (orange). All of these indicate separate introductions, though some new sequences link together to form tight clusters. This could indicate local transmission or a common source.
10/18
10/18
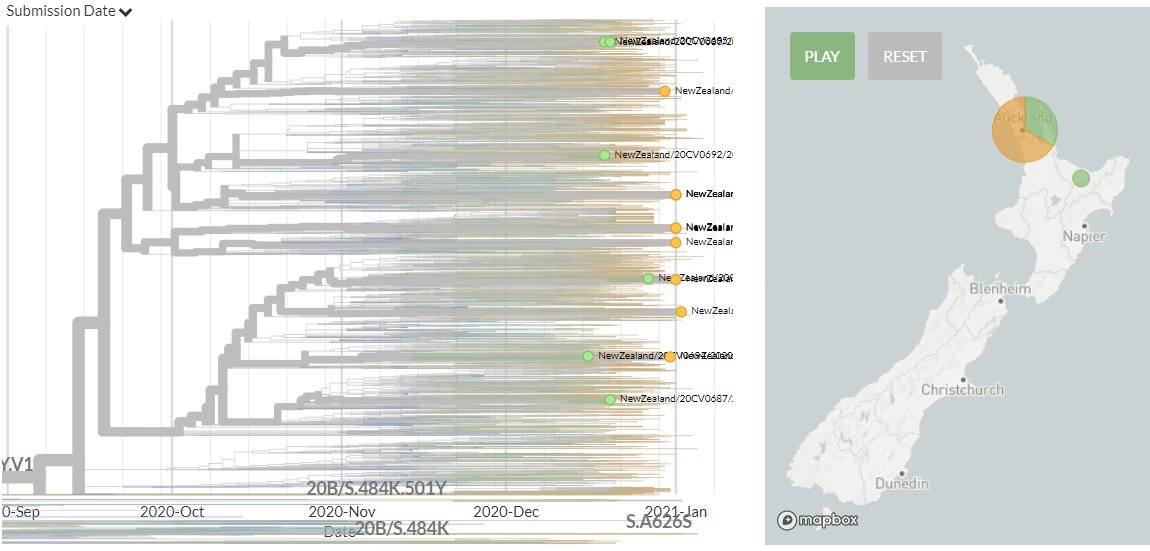
New Zealand has 10 new sequences (orange). All represent separate introductions, though some new sequences are linked together.
11/18
11/18
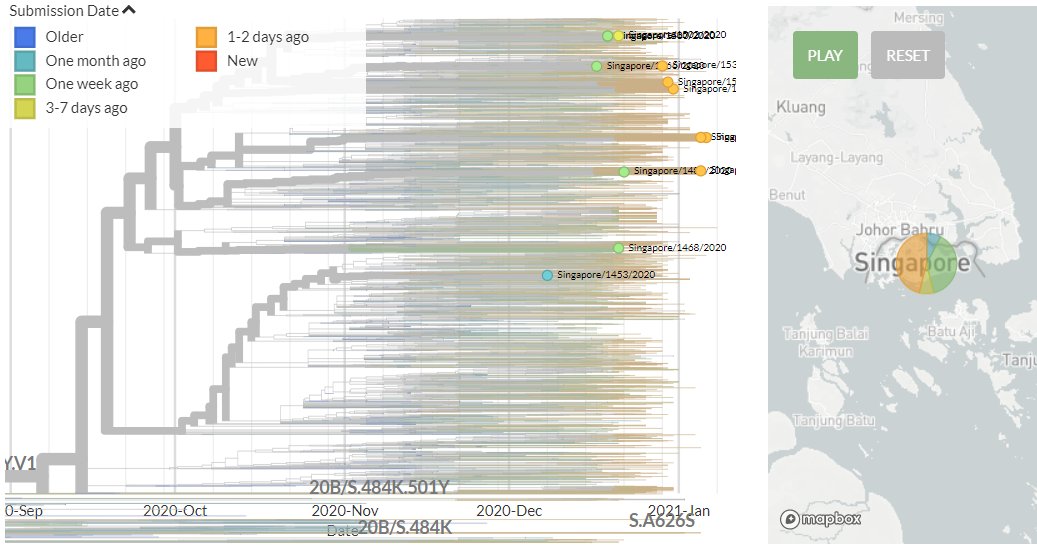
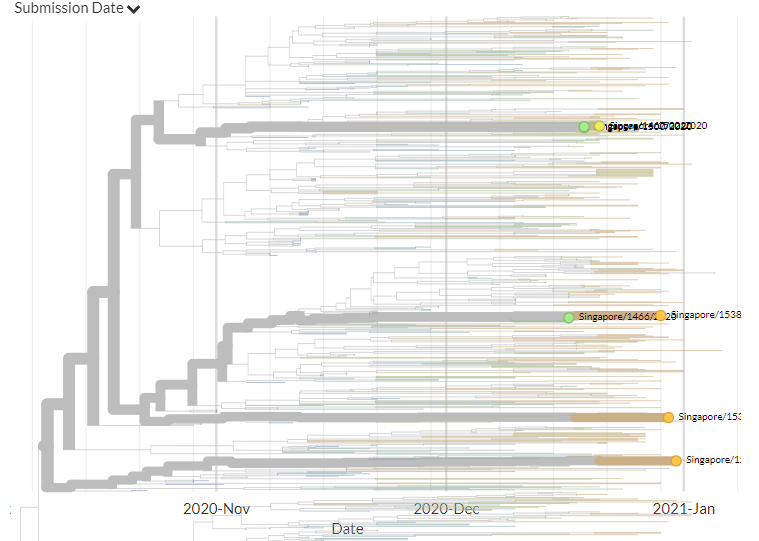
Singapore has 6 new sequences (orange). Though hard to see in zoomed-out view, most of these indicate separate introductions.
12/18
12/18
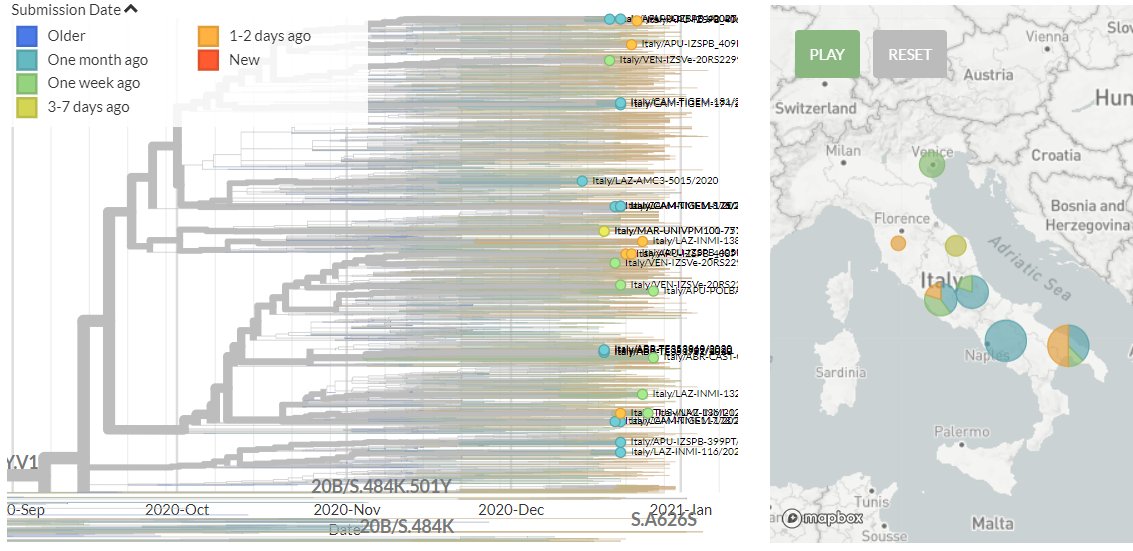
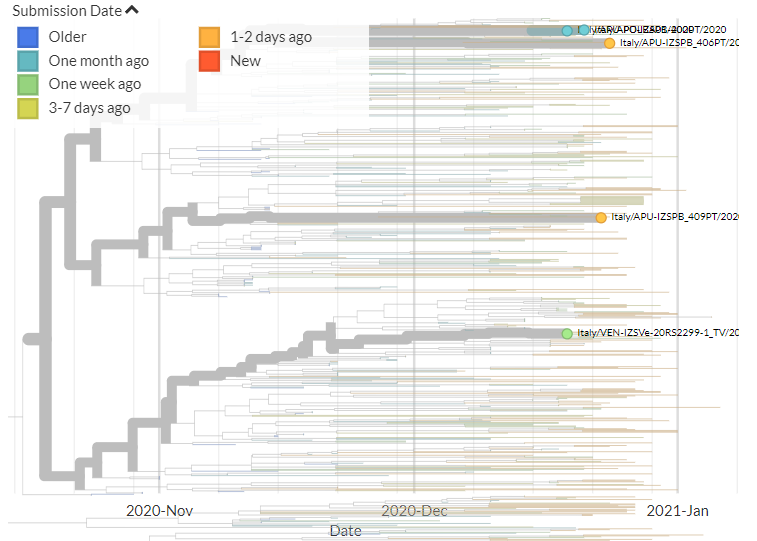
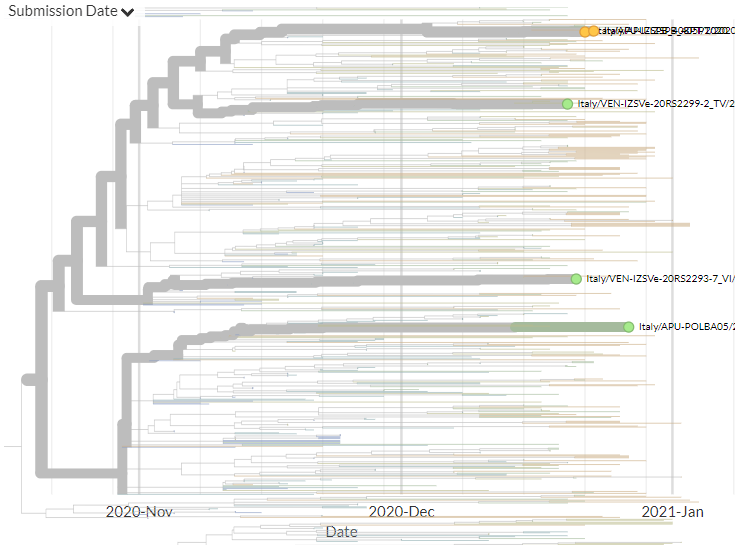
Italy has 6 new sequences (orange). It's a bit hard to see in the zoomed-out view, but none of these link directly with older samples, indicating separate introductions - though 2 new seqs link together.
13/18
13/18
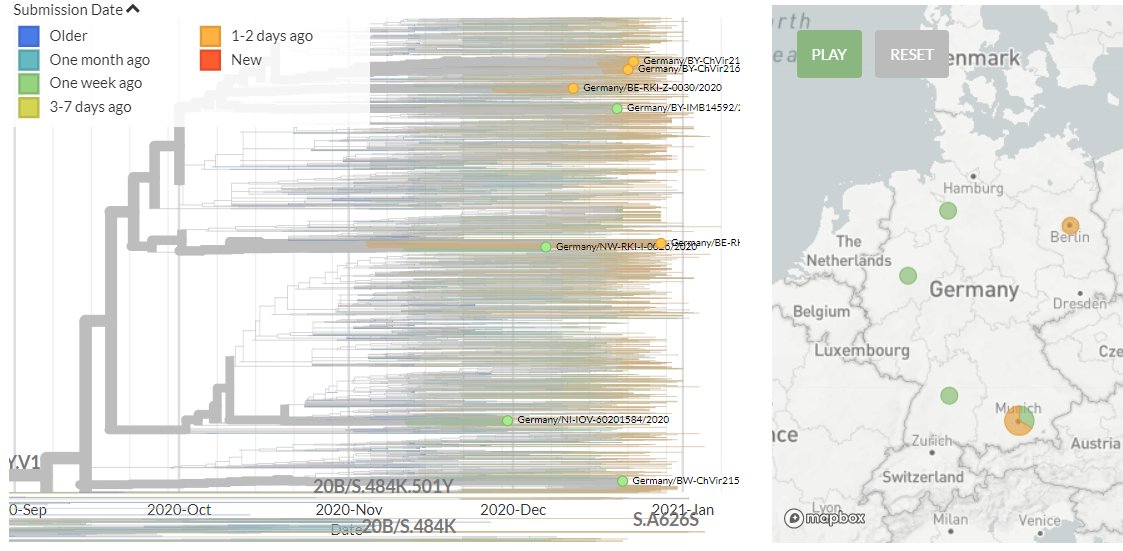
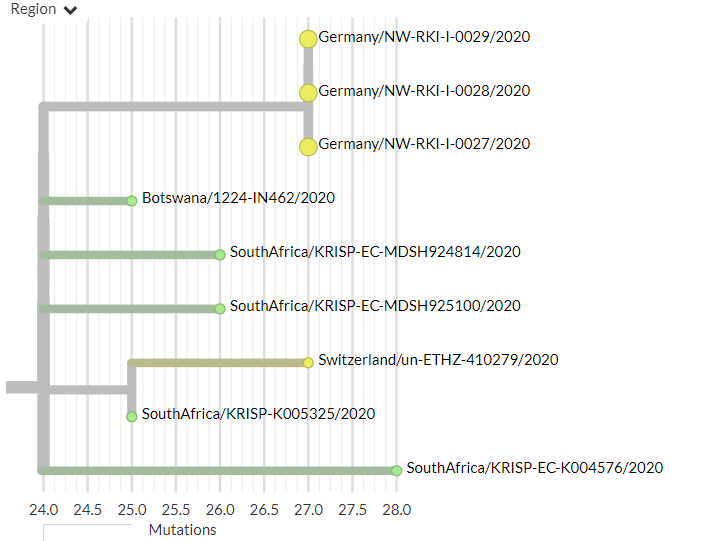
Germany has 4 new sequences (orange). Though hard to see in the zoomed-out view, these all represent separate introductions.
14/18
14/18
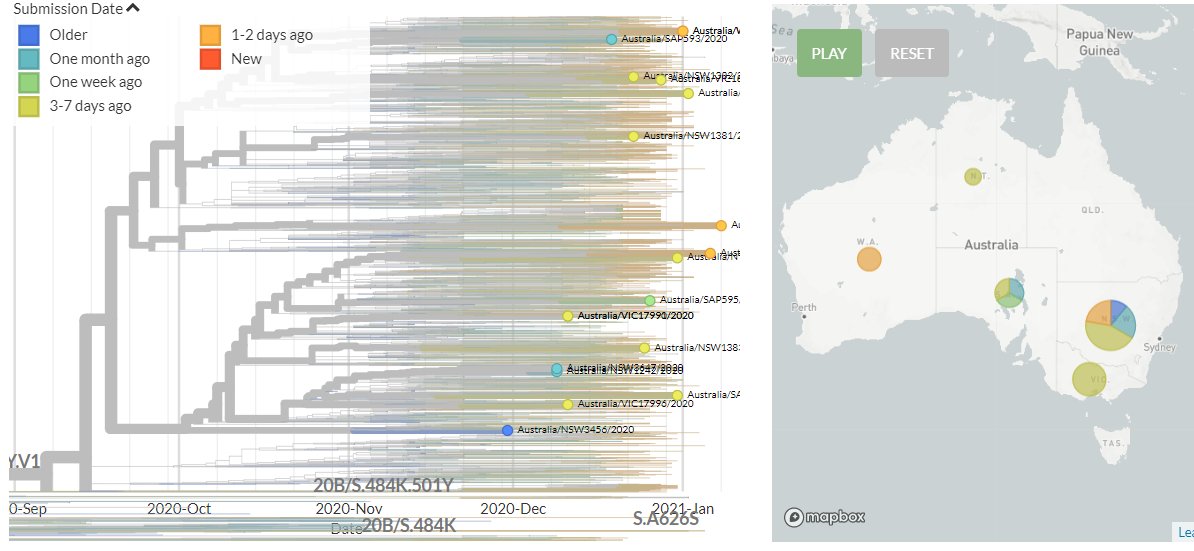
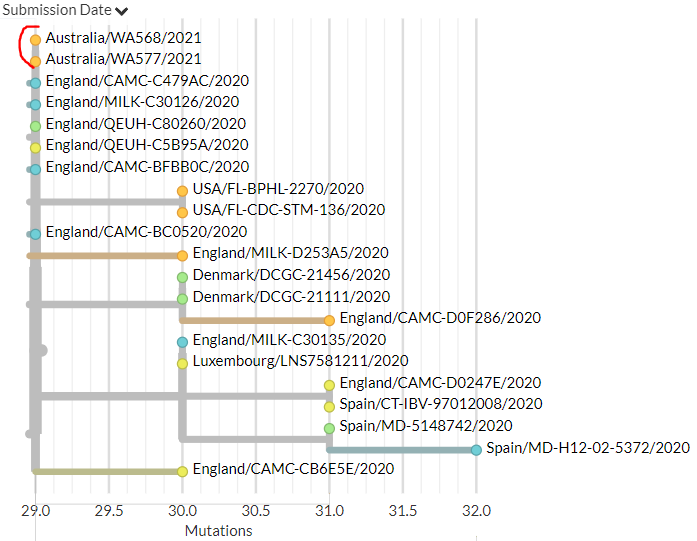
Australia also has 4 new sequences (orange), including the first from Western Australia. These indicate separate introductions, though 2 of the new sequences are identical (marked in red).
15/18
15/18
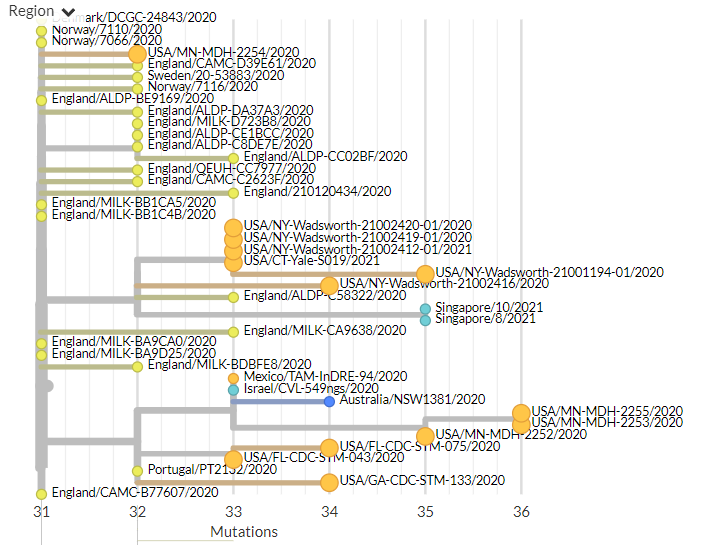
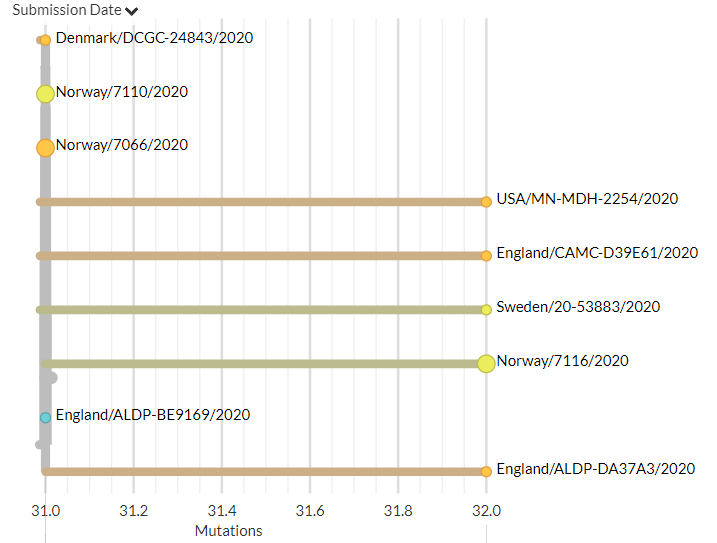
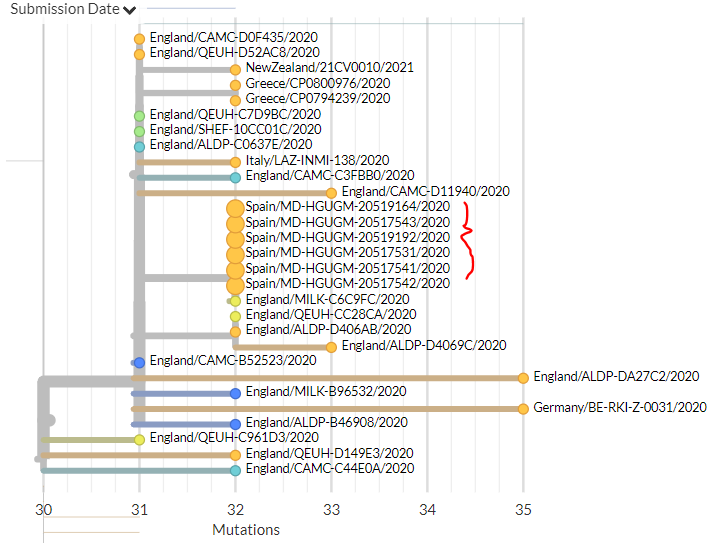
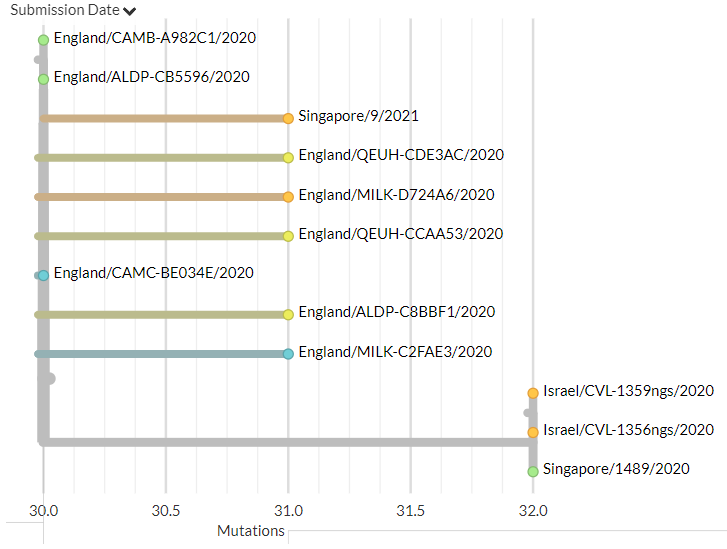
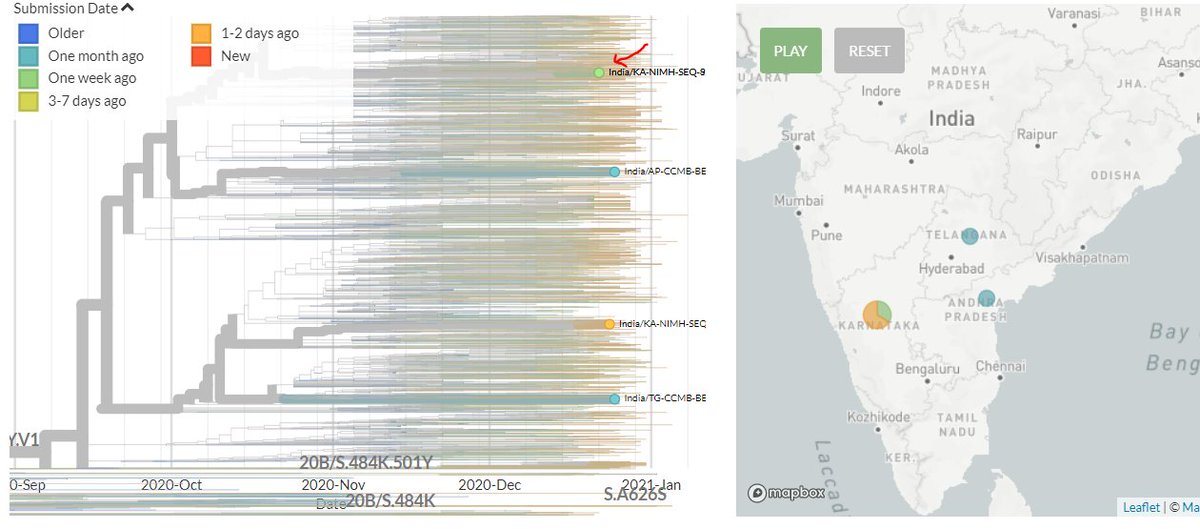
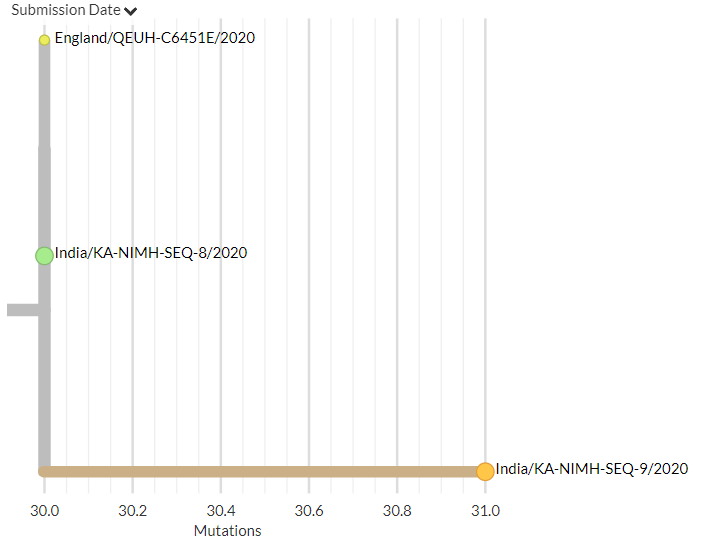
India has 2 new sequences (orange, one hidden behind the green - pic 1). One is a separate introduction, the other links with a previous sequence (pic 2).
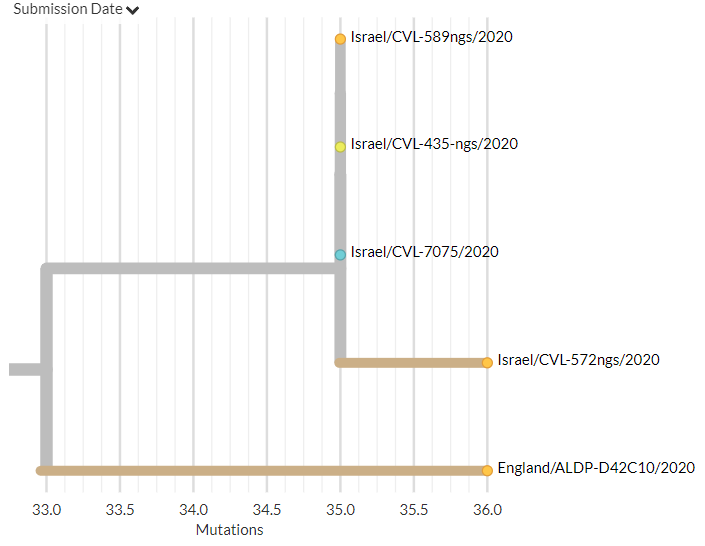
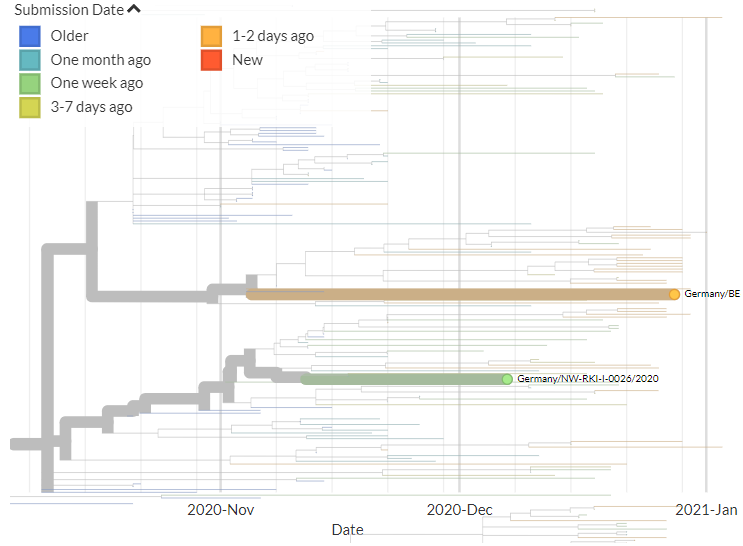
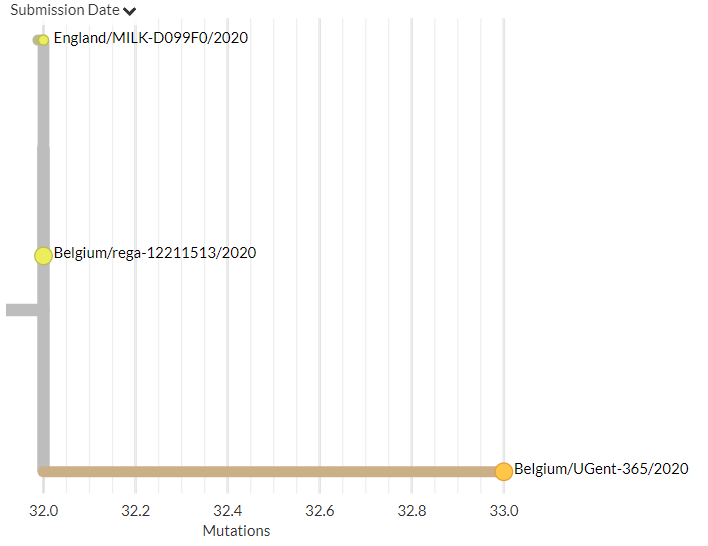
Belgium has 1 new sequence, which links 1 mutation away from a previous sequence (pic 3).
16/18
Belgium has 1 new sequence, which links 1 mutation away from a previous sequence (pic 3).
16/18
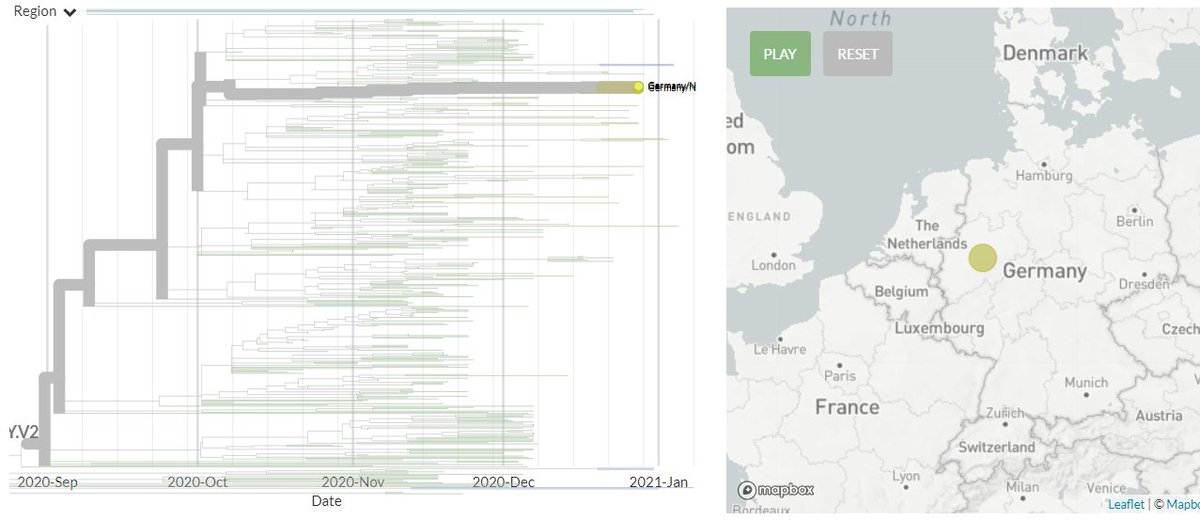
There are 3 new non-South African sequences in 501Y.V2, from Germany for the first time. They are identical, which may indicate a common exposure.
17/18
17/18
The updated country plots will go up soon, and I'll add another thread later on S:E484 & the 'Ohio variants'.
18/18
18/18
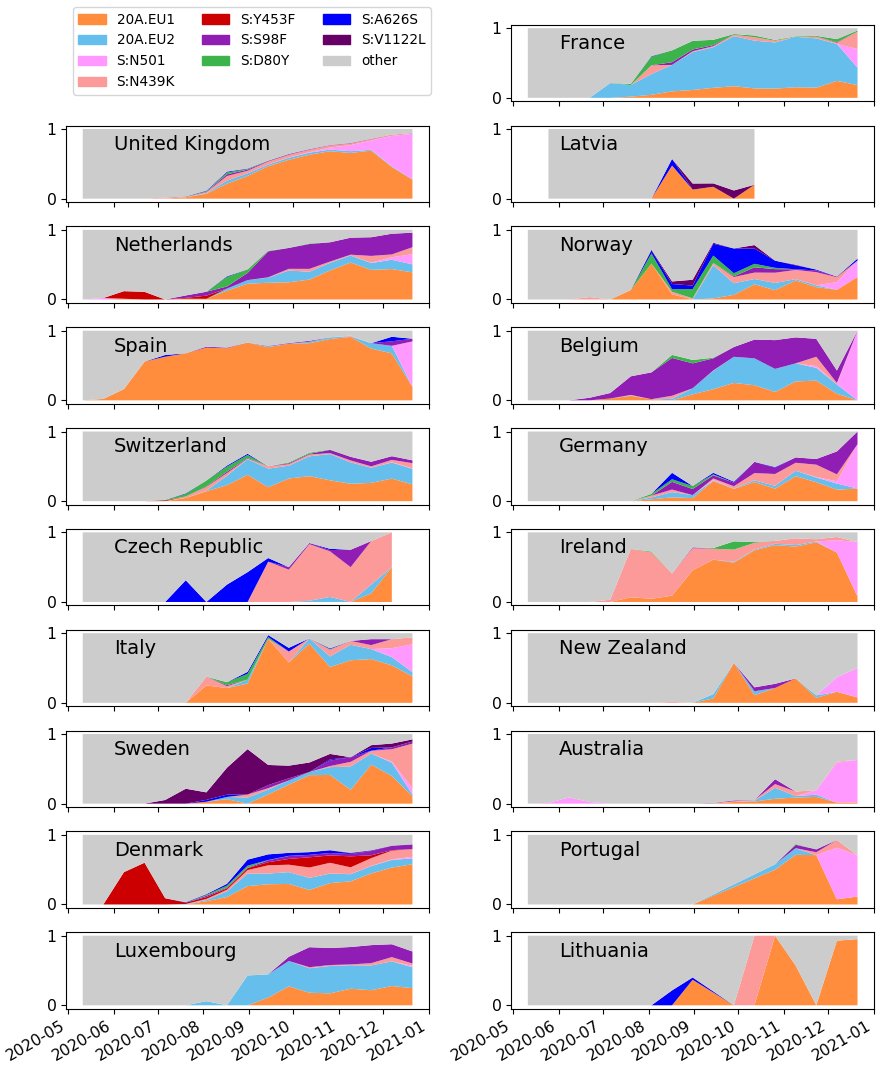
The updated country plots are now up. As always, be careful interpreting plots as many countries are selectively sequencing S:N501 & S-drop outs (which often increases S:N439), so frequencies are often not representative!
19/18
https://github.com/hodcroftlab/covariants/blob/master/country_overview.md
19/18
https://github.com/hodcroftlab/covariants/blob/master/country_overview.md

 Read on Twitter
Read on Twitter