1\\But but... you ran it to 40-45 cycles?
This is an argument that needs dissection.
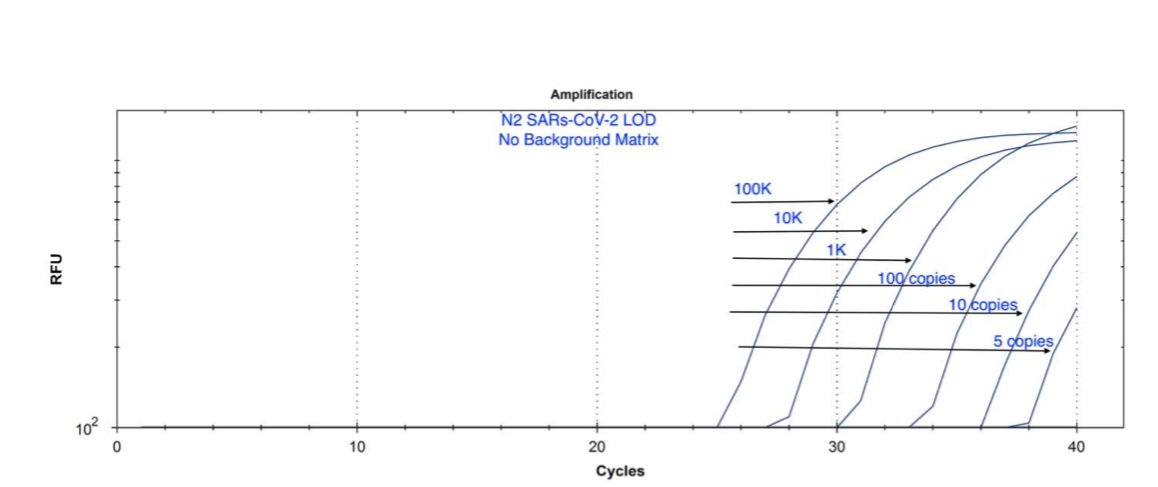
The protocols you see in EUAs are speaking to the Camera time not the Call time. To call at 37Ct... you need to see the slope of the curve out to 40. It doesn't mean they are calling at 40.
This is an argument that needs dissection.
The protocols you see in EUAs are speaking to the Camera time not the Call time. To call at 37Ct... you need to see the slope of the curve out to 40. It doesn't mean they are calling at 40.
2/Summertime positivity rates cannot be used as a proxy for wintertime false positive rates.
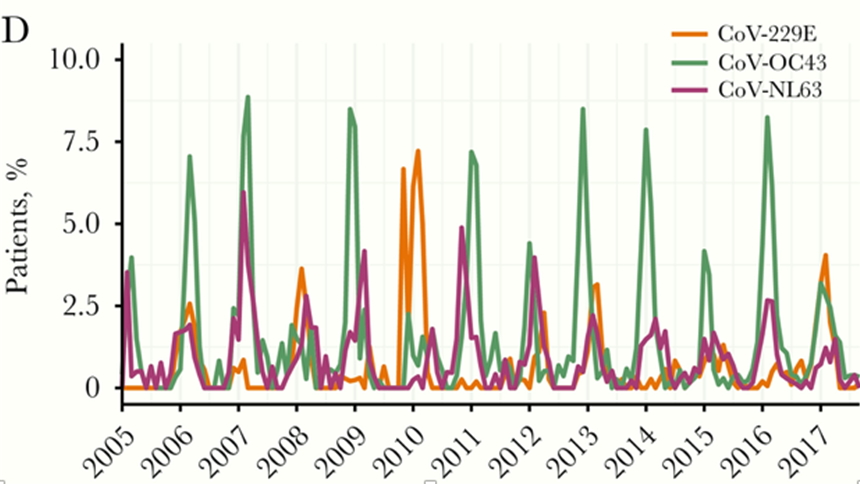
The virome in the winter is rich in HCoVs not present in the summer. I would assume the background microbiome is as well.
The virome in the winter is rich in HCoVs not present in the summer. I would assume the background microbiome is as well.
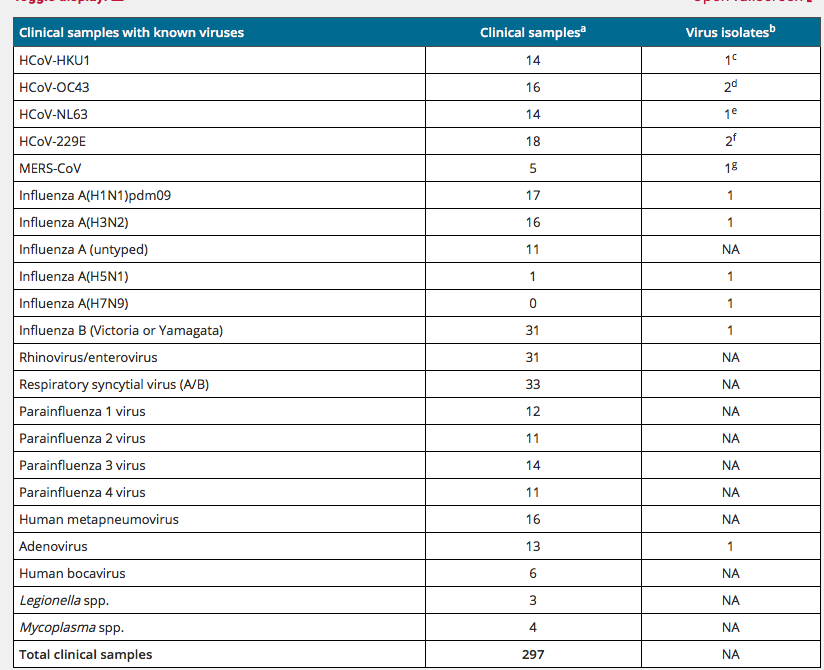
But we tested the Primers against HCoV in our exclusion analysis!
Sure.... but Just 4.
We just witness the S-gene amplicon drop out in the second season of C19 and we should expect HCoVs to have similar diversity.
Sure.... but Just 4.
We just witness the S-gene amplicon drop out in the second season of C19 and we should expect HCoVs to have similar diversity.
3/But we sequence a subset of samples.
Squid ink. Is the data public?
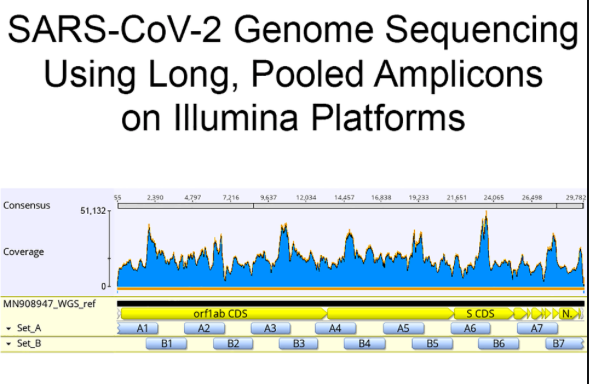
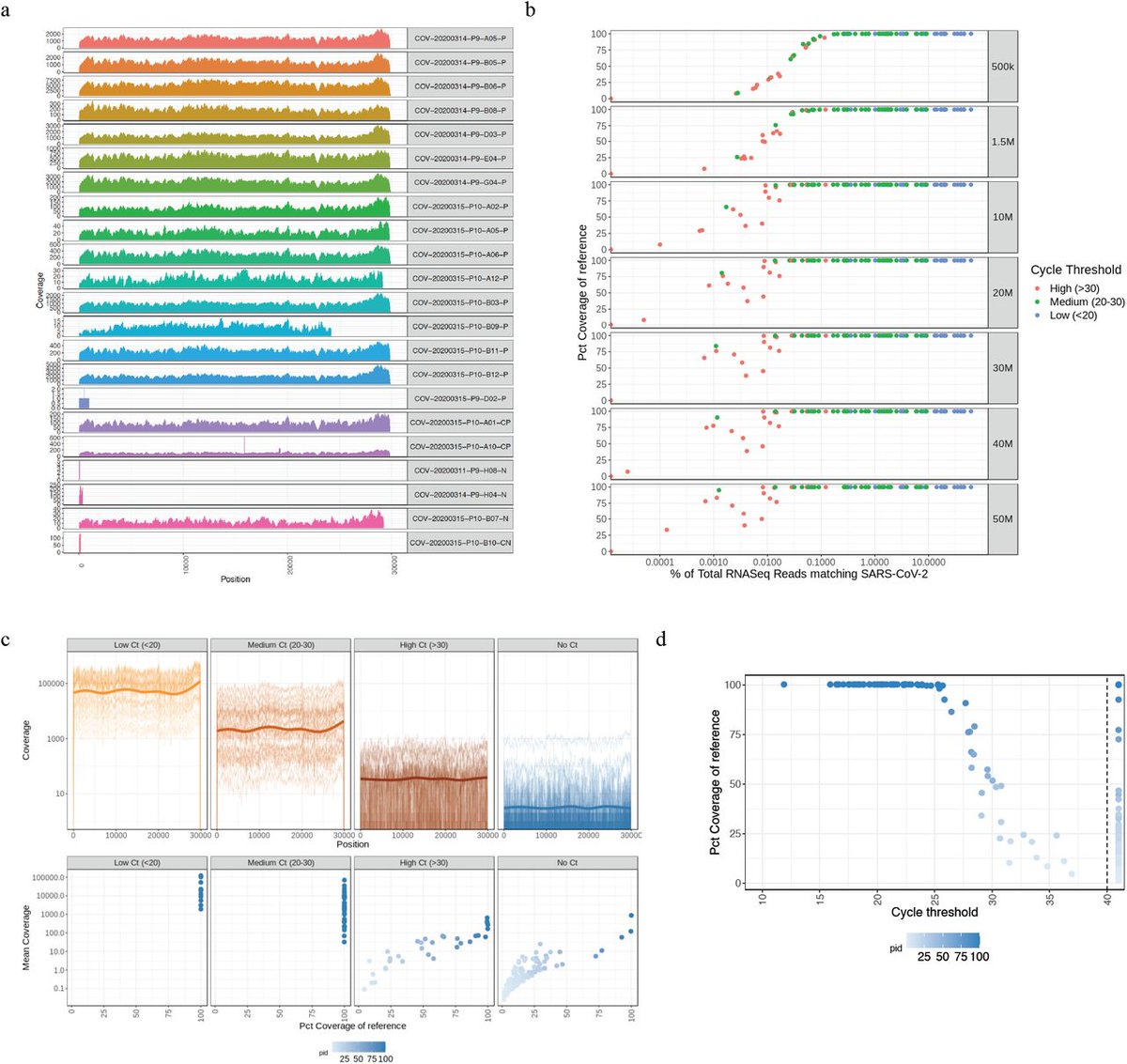
Sequencing is usually performed on samples with low Ct as its difficult/^$ to sequence samples with high Ct.
Many sequencing methods must amplify the genome before sequencing it.
Squid ink. Is the data public?
Sequencing is usually performed on samples with low Ct as its difficult/^$ to sequence samples with high Ct.
Many sequencing methods must amplify the genome before sequencing it.
So lets say you have a sample that has more HCoV than SARs. You go to PCR its genome with a couple dozen different primers than the primers that delivered you the qPCR signal. You are going to sequence what amplifies most efficiently. This is not a whole virome survey. This is..
Mason's lab has done this on 200+ samples but this is not what is in routine use for SARs Sequence confirmation (I hope to be proven wrong here overtime).
The PCR for sequencing is run to a set number of cycles usually below 30. https://www.biorxiv.org/content/10.1101/2020.04.20.048066v5
The PCR for sequencing is run to a set number of cycles usually below 30. https://www.biorxiv.org/content/10.1101/2020.04.20.048066v5
And it's performed on samples that qPCR below a certain threshold so there are multiple selections going on with what is being sequence confirmed.
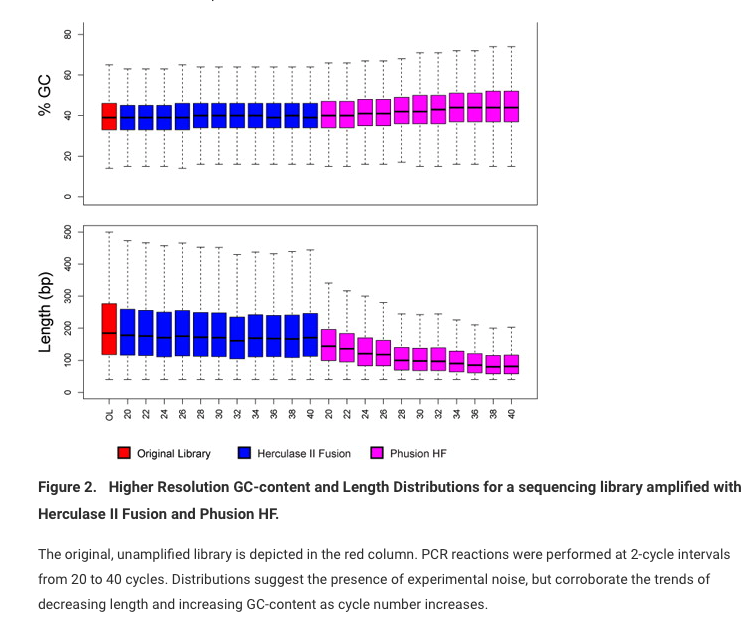
When PCR cycles are used prior to sequencing you loose quantitation information in the sequencing data.
https://www.future-science.com/doi/10.2144/000113809
When PCR cycles are used prior to sequencing you loose quantitation information in the sequencing data.
https://www.future-science.com/doi/10.2144/000113809
All that said, I think the larger source for FPs is related to how long people remain qPCR positive post infectiousness.
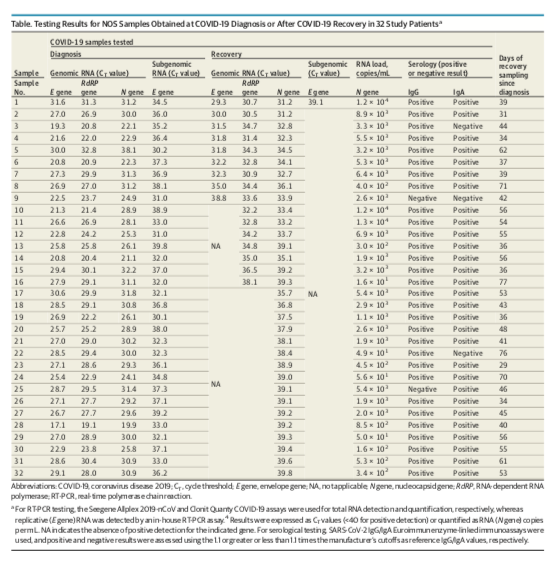
The infection window is 7-10 days and Liotti et al has seen patients 77 days qPCR positive (mean 48.6). So there could be a 5:1 - 10:1 false quarantine rate.
The infection window is 7-10 days and Liotti et al has seen patients 77 days qPCR positive (mean 48.6). So there could be a 5:1 - 10:1 false quarantine rate.
Labs will claim there is nothing they can do about this. Its not a FP as we picked up RNA.
They could stick to testing only symptomatics to improve their PPV and test twice to get a viral load change.
They could stick to testing only symptomatics to improve their PPV and test twice to get a viral load change.
But But.. The real problem is False negatives so we cant afford to cut the cycle number.
This is not true. The false negatives are from an entirely different problem related to the sample prep and swabbing procedures being highly variable. No amount of PCR will rescue Poisson.
This is not true. The false negatives are from an entirely different problem related to the sample prep and swabbing procedures being highly variable. No amount of PCR will rescue Poisson.
Dahdouh et al cover this.
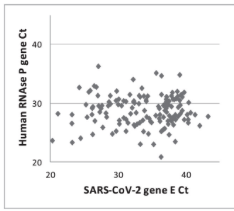
10-13 Ct variance in Human RNaseP gene. This is a matrix control that tells you how well your collection and prep performed. If you do not normalize to this you cannot honestly call it a viral load as you have no denominator. https://pubmed.ncbi.nlm.nih.gov/33131699/
10-13 Ct variance in Human RNaseP gene. This is a matrix control that tells you how well your collection and prep performed. If you do not normalize to this you cannot honestly call it a viral load as you have no denominator. https://pubmed.ncbi.nlm.nih.gov/33131699/
One of our complaints about the Corman-Drosten test is that is has no Human RNaseP gene target so you cant actually measure viral load.
These sampling False negatives should not be conflated with the false negatives due to poor primer design and PCR sensitivity. They are diff.
These sampling False negatives should not be conflated with the false negatives due to poor primer design and PCR sensitivity. They are diff.
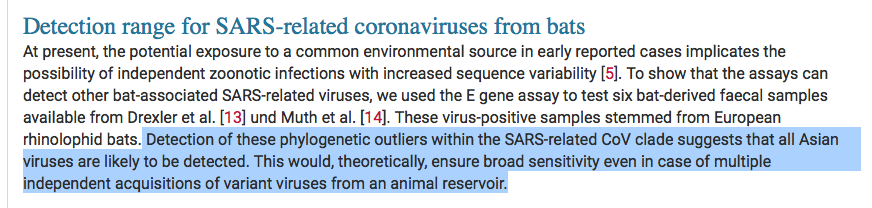
So the 'UK scariant' is a reminder that our EUAs are built on as little as 40 samples in the test set with a limited diversity in the exclusion test. The sequencing assays being used to claim specificity have their own bias. But the long tail of +vity is whats killing us.

 Read on Twitter
Read on Twitter