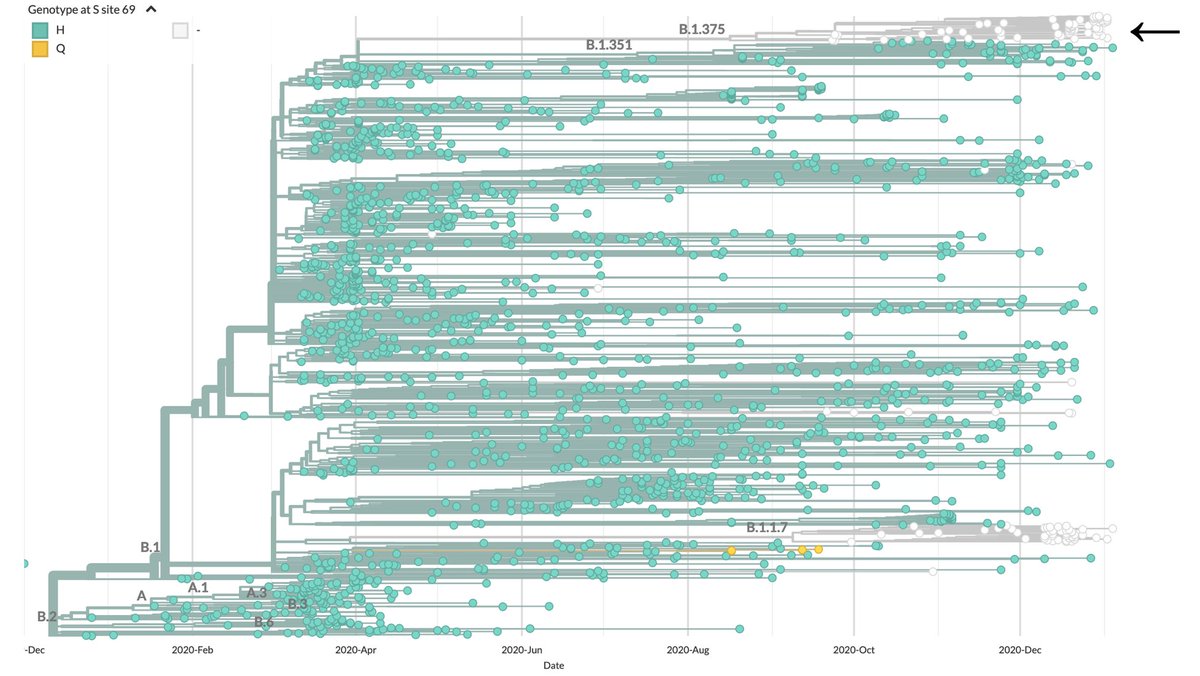
The amino acid deletion ∆69/70 has been used as a proxy for detecting viruses from B.1.1.7 ('UK variant'). The same marker has been used in the US, but we caution against its use. We identified a new SARS-CoV-2 lineage (B.1.375) with that same feature. https://virological.org/t/detection-of-non-b-1-1-7-spike-69-70-sequences-b-1-375-in-the-united-states/587
A new lineage circulating in the US (B.1.375, arrow) shows the same deletion ∆69/70 in the spike protein, and a set of unique amino acid substitutions were found in this lineage: ORF1a T1828A, ORF1b E1264D, ORF3a T151I, and M I48V.
Viruses in the lineage B.1.375 have been identified in all regions of the US, specially in states where genomic surveillance of SARS-CoV-2 has been done frequently, such as: Connecticut, Florida, California and Wisconsin.
Selecting samples with S gene target failure in TaqPath COVID-19 assays (ThermoFisher) is not a good proxy for detecting B.1.1.7 in the US, given B.1.375.
RT-PCR results alone, should not be used to infer the incidence of COVID cases caused by B.1.1.7, at least in the US.
RT-PCR results alone, should not be used to infer the incidence of COVID cases caused by B.1.1.7, at least in the US.
We don't have information about how common viruses from B.1.375 are, and so far NO data suggest that variants from this lineage are more transmissible or cause more severe illness, or increased risk of death. More research is needed. 

This report was a result of a joint effort from members of research groups in three US states:
AZ: @bblarsen1, Mike Worobey
CT: @JosephFauver @tdalpert @NathanGrubaugh
WI: @GageKMoreno @KATarinambraun @tcfriedrich David O'Connor
AZ: @bblarsen1, Mike Worobey
CT: @JosephFauver @tdalpert @NathanGrubaugh
WI: @GageKMoreno @KATarinambraun @tcfriedrich David O'Connor

 Read on Twitter
Read on Twitter