New review from my lab in @NatureRevCancer on the role of aneuploidy in tumorigenesis. https://www.nature.com/articles/s41568-020-00321-1?WT.mc_id=TWT_NatureRevCancer
A few key points to highlight:
- Certain chromosomal trisomies are found in more tumors than mutations in standard oncogenes like KRAS/PIK3CA/etc. (why??)
- Certain chromosomal trisomies are found in more tumors than mutations in standard oncogenes like KRAS/PIK3CA/etc. (why??)
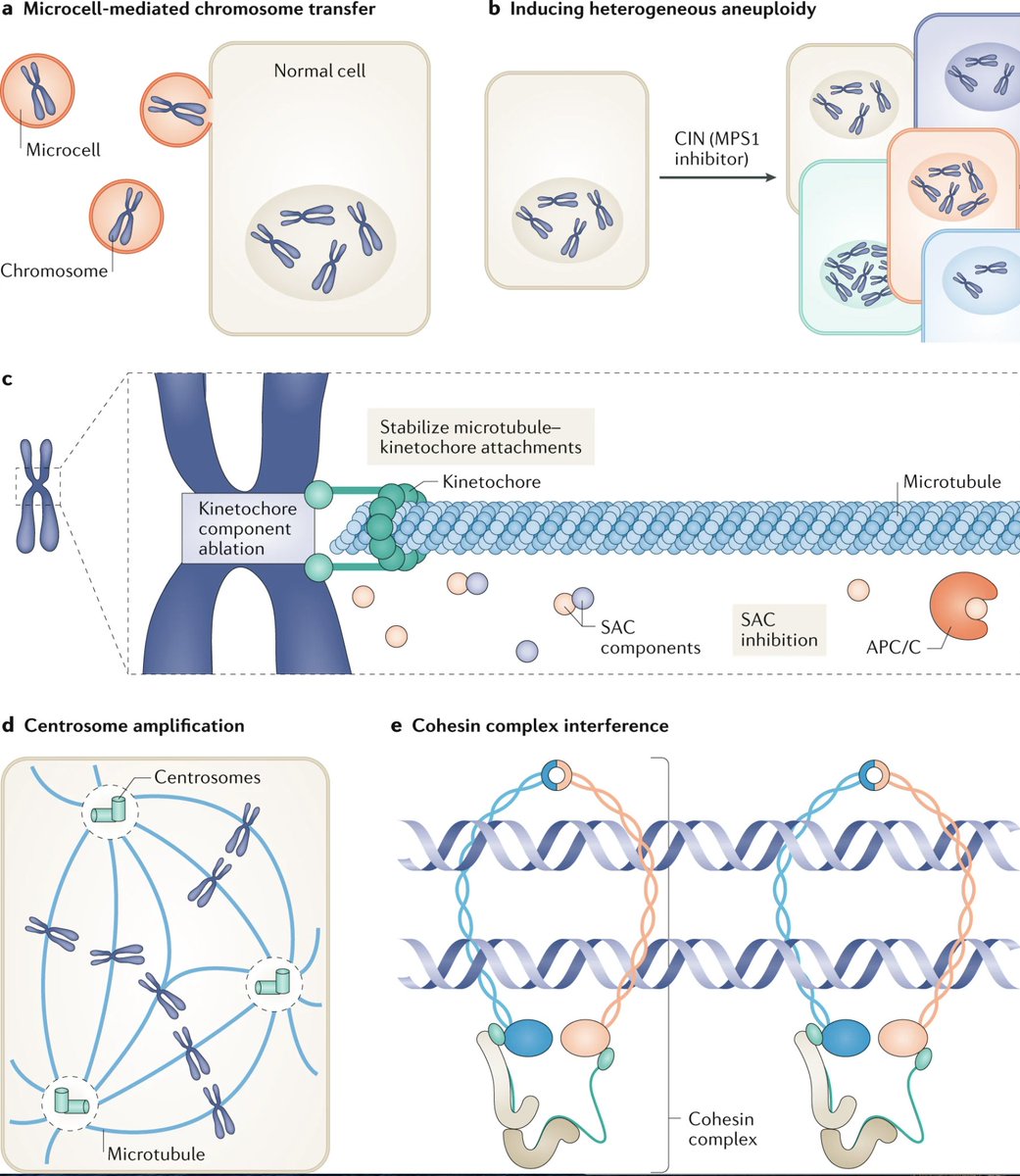
- Existing in vivo models of aneuploidy are quite limited. People have been making checkpoint-deficient mouse models for a while, but it's worth noting that these genes (BUBR1, CDC20, MAD2, etc) are almost never mutated in human tumors. Need better models!
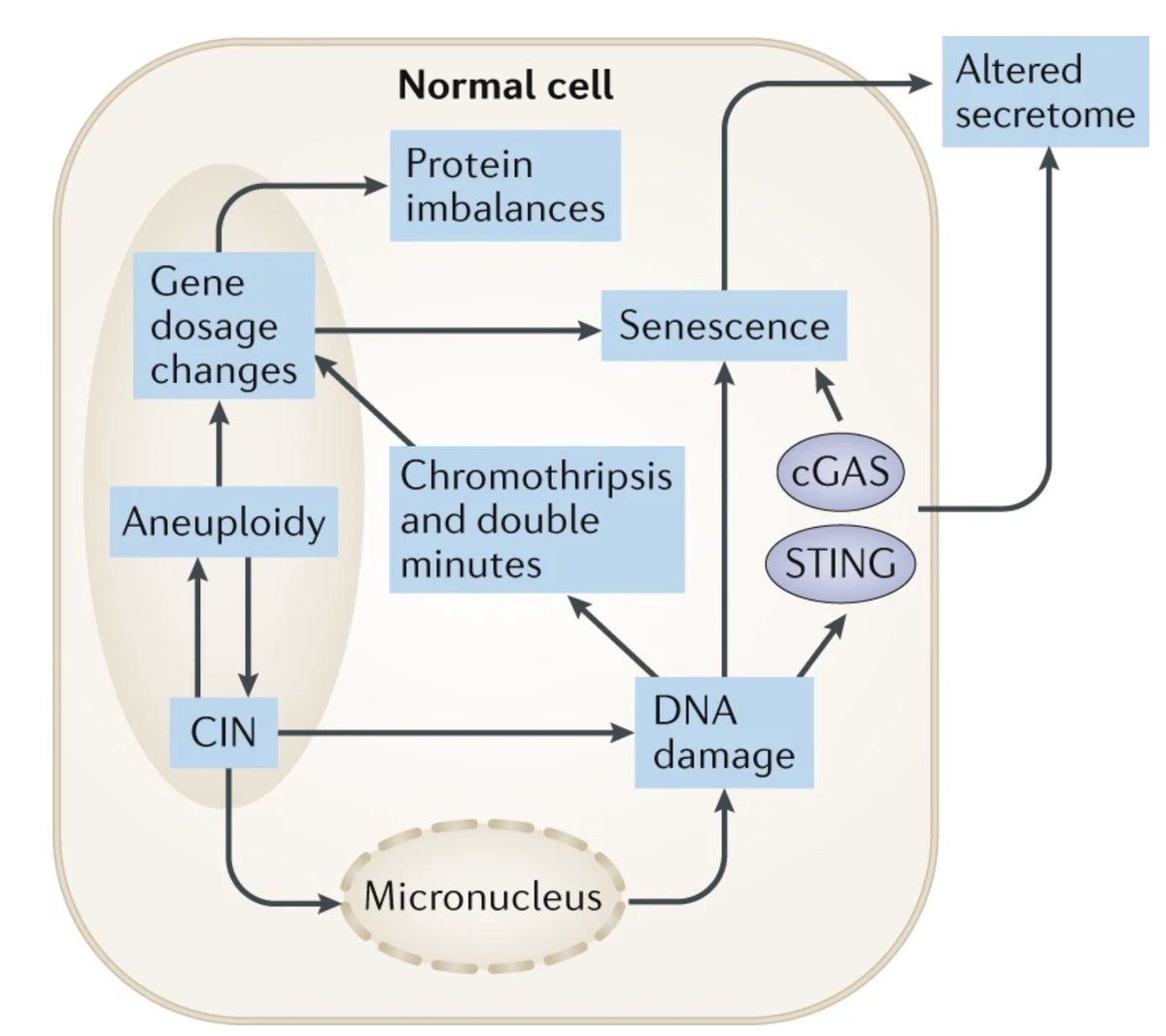
- The key driver of aneuploidy-associated phenotypes are the resulting gene dosage imbalances. Over-expressing an oncogene (like MYC in Chr8 trisomies) is clearly bad. At the same time...
Chromosomes are also physical objects, and as @PeterLyLab @EmilyHatch19 @Samuel_Bakhoum and others have shown, the physical consequences of chromosome missegregation (DNA damage/micronucleus formation/cGAS activation) are important aspects of CIN as well.
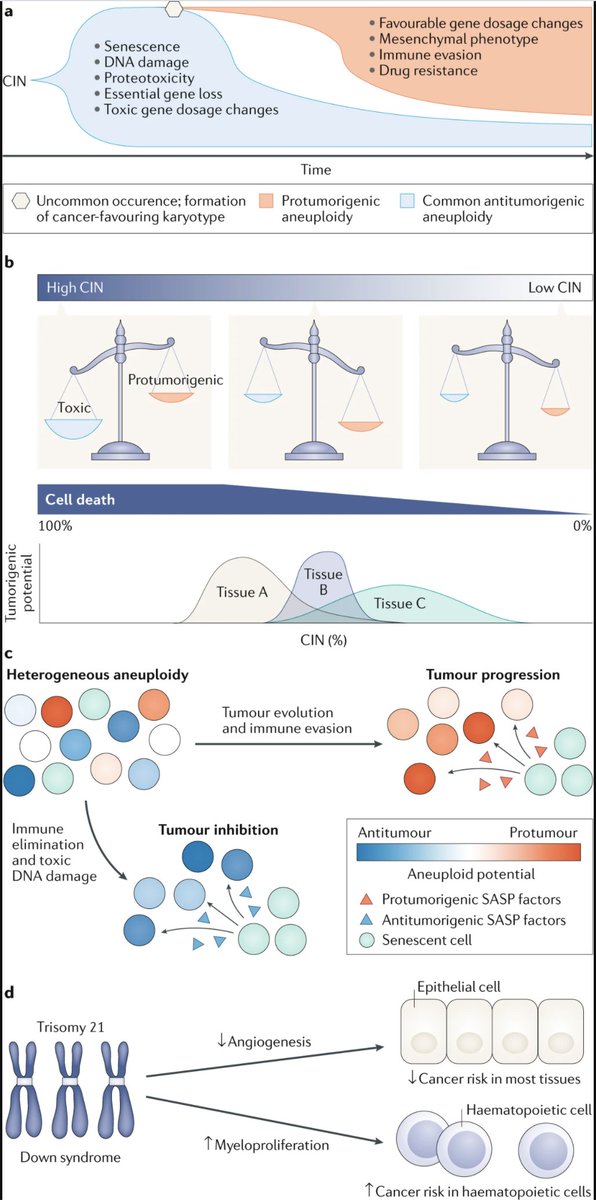
- While aneuploidy is typically associated with cancer progression, it is not a universal cancer driver. For instance, individuals with Down syndrome have higher rates of leukemia, but they also have significantly lower rates of solid tumor formation.
So a comprehensive understanding of aneuploidy should account for both its pro-tumorigenic and its tumor suppressive consequences.
Finally, in my view, aneuploidy is a very promising target for therapy development. While aneuploidy is pervasive in cancer, its extremely rare in normal tissue. Could we train the immune system to selectively eliminate cells harboring chromosome changes?? More work is needed!
Hope you enjoy the review. Any questions/feedback, please let me know!

 Read on Twitter
Read on Twitter