My 1st-1st author paper is out! There were many moments in this >four year journey where I honestly doubted we would ever publish. Let me tell you a bit about the paper, which should maybe be called "how NOT to measure the S isotopes of amino acids" …https://onlinelibrary-wiley-com.clsproxy.library.caltech.edu/doi/abs/10.1002/rcm.9007
Amino acids have been measured for C, N, O, even H isotopes - but nobody had yet tackled S. At the start of my PhD my advisor and I thought, sure, let's try it, can't be too hard right?
Narrator: It was really hard.
Narrator: It was really hard.
Why is measuring sulfur amino acids (cysteine and methionine) so hard? They react with oxygen, forming numerous products. For sulfur isotopes, that was not going to be okay - we had to either keep them reduced or push them to be oxidized. We chose reduced (we were wrong).
Another problem was on the instrument end: sulfur isotope analysis by EA-IRMS relies on combustion to SO2 - a polar gas that sticks to columns, making peaks wide and short. This lowers signal to noise ratios, which means you need more sample for a good peak.
After two years of failing to prevent oxidation reactions throughout our workup, we switched tactics, looking to quantitatively oxidize cys to cysteic acid and met to methionine sulfone. After some trial and error, we settled on performic acid as an oxidant. Our new method:
For the instruments end, we were lucky enough to test a new @thermofisher EA-IRMS that has programmable flow rates and a ramped GC column - this allows you to trap that sticky SO2 gas at low temp, then release it later at lower flow rates and higher temps -now: nice, tall peaks!
This new design was a spectacular improvement, allowing us to measure organic and inorganic sulfur between 1 and 10 µg S at <0.3‰ precision. Our previous EA-IRMS required 100s of µgs.
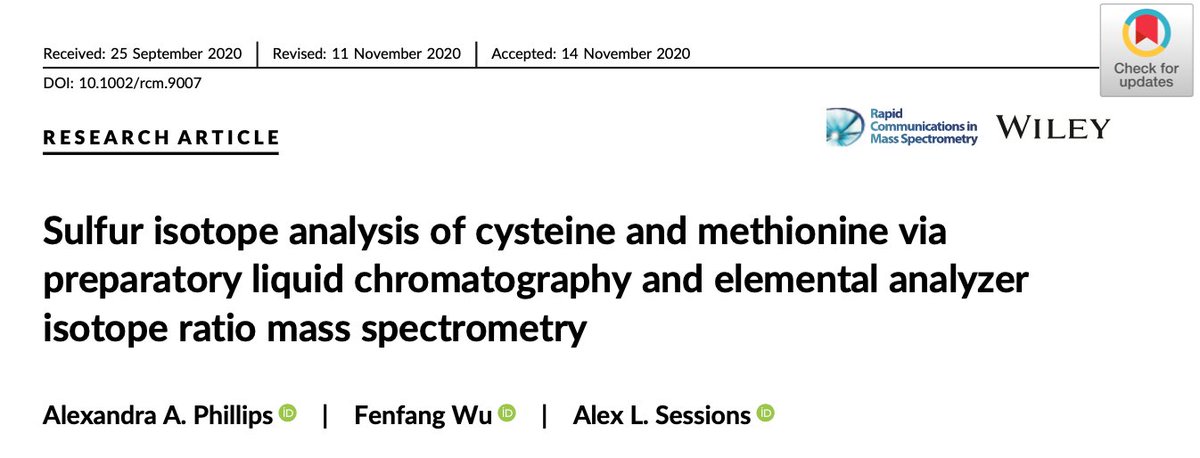
This really opened the window for analyzing cys and met from natural samples. Over more years of other methods development (with many late nights crying at the HPLC) I managed to measure the sulfur isotopes of four organisms: two fish species and two bacteria.
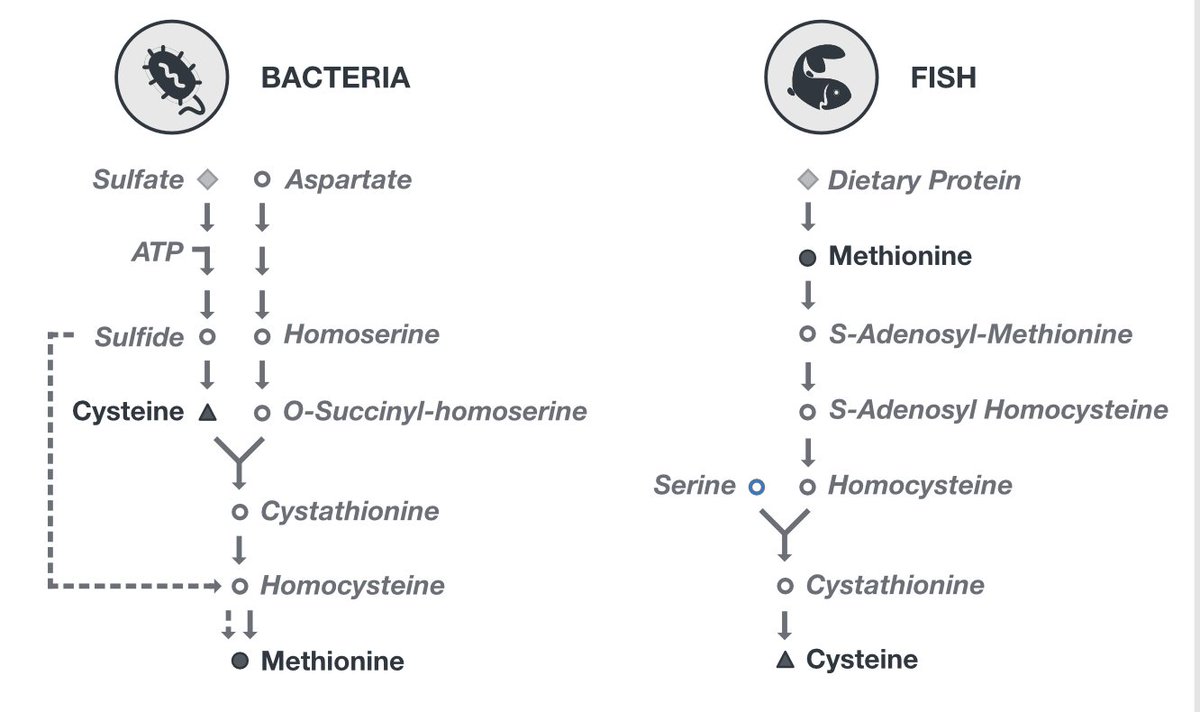
Let's zoom in on E coli and T albacares (tuna). In E coli, cys is enriched in 34S compared to met - but for T albacares this pattern is reversed. We think this is a direct reflection of metabolism (+ KIEs), as bacteria synthesize met from cys while fish make cys from dietary met.
This direct link to metabolism is really promising for ecological studies of both modern and ancient environments - and hopefully highlights the improved clarity isotope geochemists can get with compound specific work like this.
I'm really excited this work is out there in the world, and hope that others either improve on this method or apply it to some interesting new problems! Thank you to all of my friends in and out of the lab who helped me get through this difficult, but rewarding project.

 Read on Twitter
Read on Twitter