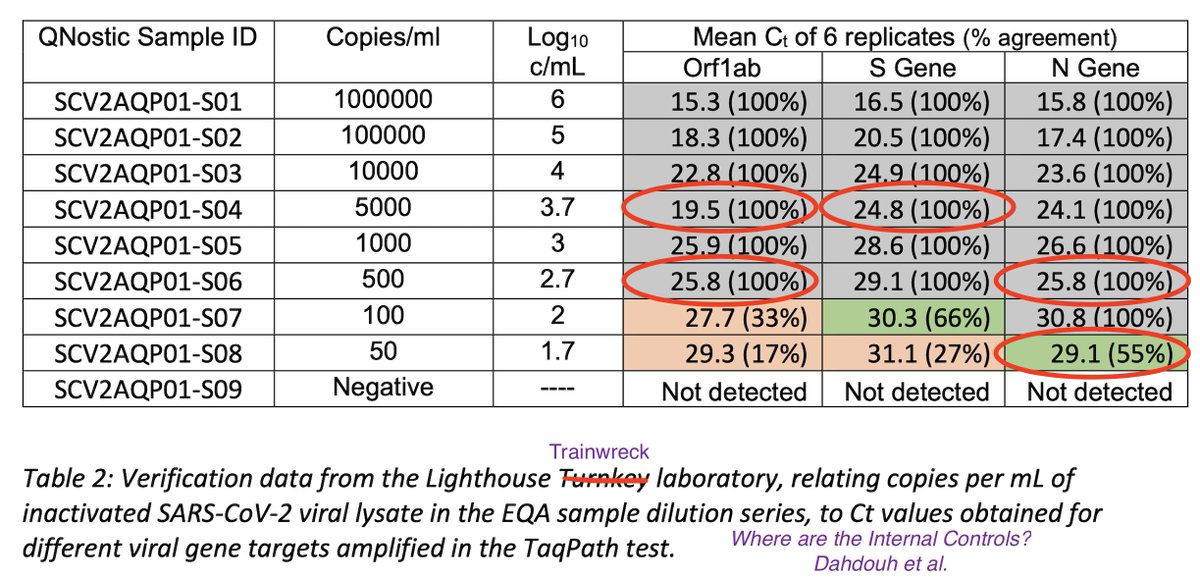
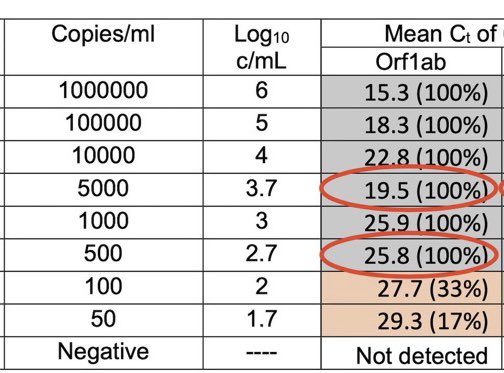
If you are in the UK, this is the serial dilution of the test being run on you for C19.
For each 10 fold drop in Copies/ml, you should see a 3.3 increment in Ct.
What do you see?
Why does a 5000 copies/ml sample have a lower Ct than a 10,000 cp/ml sample?
For each 10 fold drop in Copies/ml, you should see a 3.3 increment in Ct.
What do you see?
Why does a 5000 copies/ml sample have a lower Ct than a 10,000 cp/ml sample?
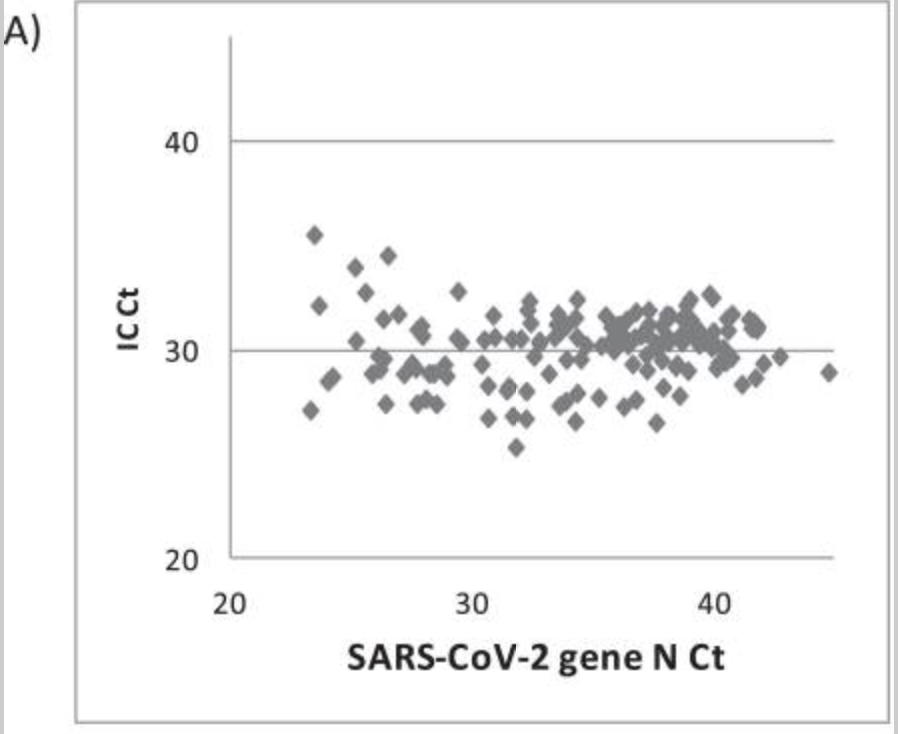
The paper does speak to a spike in MS-2 control but this isn't a true internal control like RNaseP.
Spike-in internal controls are important for measuring assay drop out. They prove the PCR was functional.
But RNaseP controls which amplify human DNA/RNA harvested on the Swab..
Spike-in internal controls are important for measuring assay drop out. They prove the PCR was functional.
But RNaseP controls which amplify human DNA/RNA harvested on the Swab..
These ICs are more important as you really cant quantitate viral load and make claims about S gene knock outs being higher copy number if you don't normalize the Swab collection and sample prep issues. These can vary 10-16 Cts according to Dahdouh et al.
https://www.medrxiv.org/content/10.1101/2020.12.24.20248834v1.full.pdf
https://www.medrxiv.org/content/10.1101/2020.12.24.20248834v1.full.pdf
The paper omits the MS-2 Cts and doesnt really address Dahdouh et al concerns regarding the large Ct variance in swab to swab sample prep.
If you don't normalize for the sample prep variance, and go on to make panic inducing statements about viral load...
you are a hack.
But given the serial dilutions they just published... we already knew this.
you are a hack.
But given the serial dilutions they just published... we already knew this.

 Read on Twitter
Read on Twitter