After many months of hard work our analysis of the lineage structure and importation dynamics of the first wave of the #SARSCoV2 epidemic in the UK is out in @ScienceMagazine today! https://science.sciencemag.org/content/early/2021/01/07/science.abf2946
begin thread
begin thread
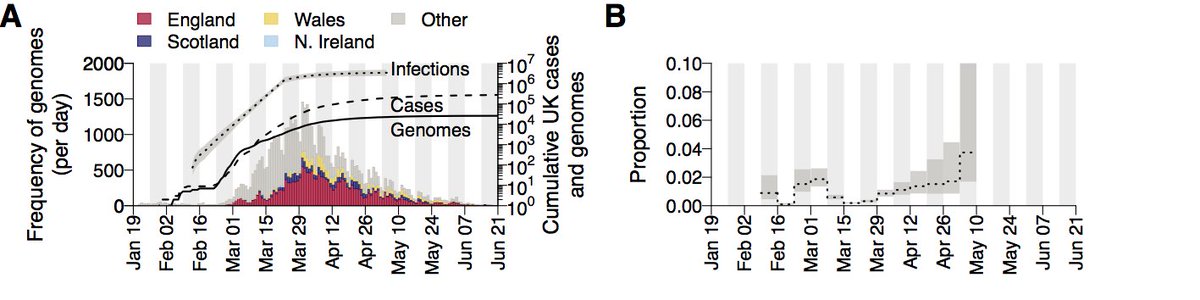
We use >50K genomes, 26K from the UK (thanks to @CovidGenomicsUK) representing ~10% of UK cases in the 1st wave, but still only ~0.5% of all estimated infections. Nonetheless this is a level of surveillance much higher than we’re used to and allows for unprecedented insights.
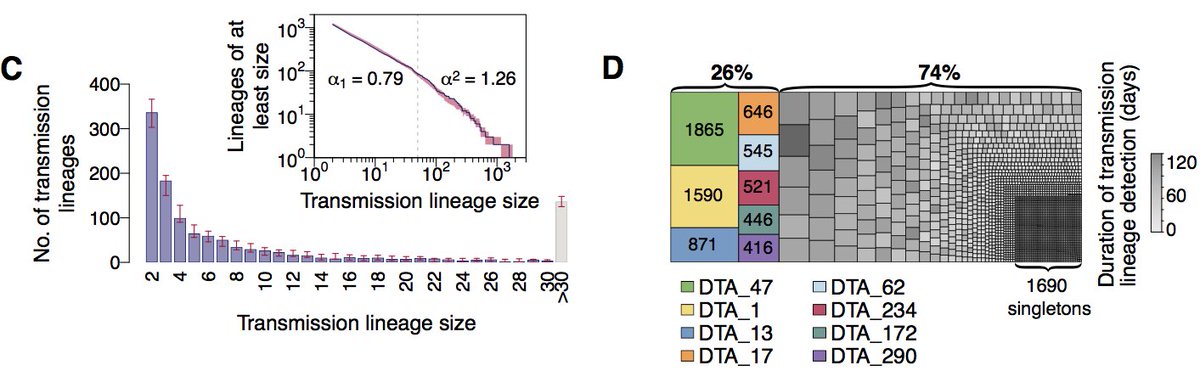
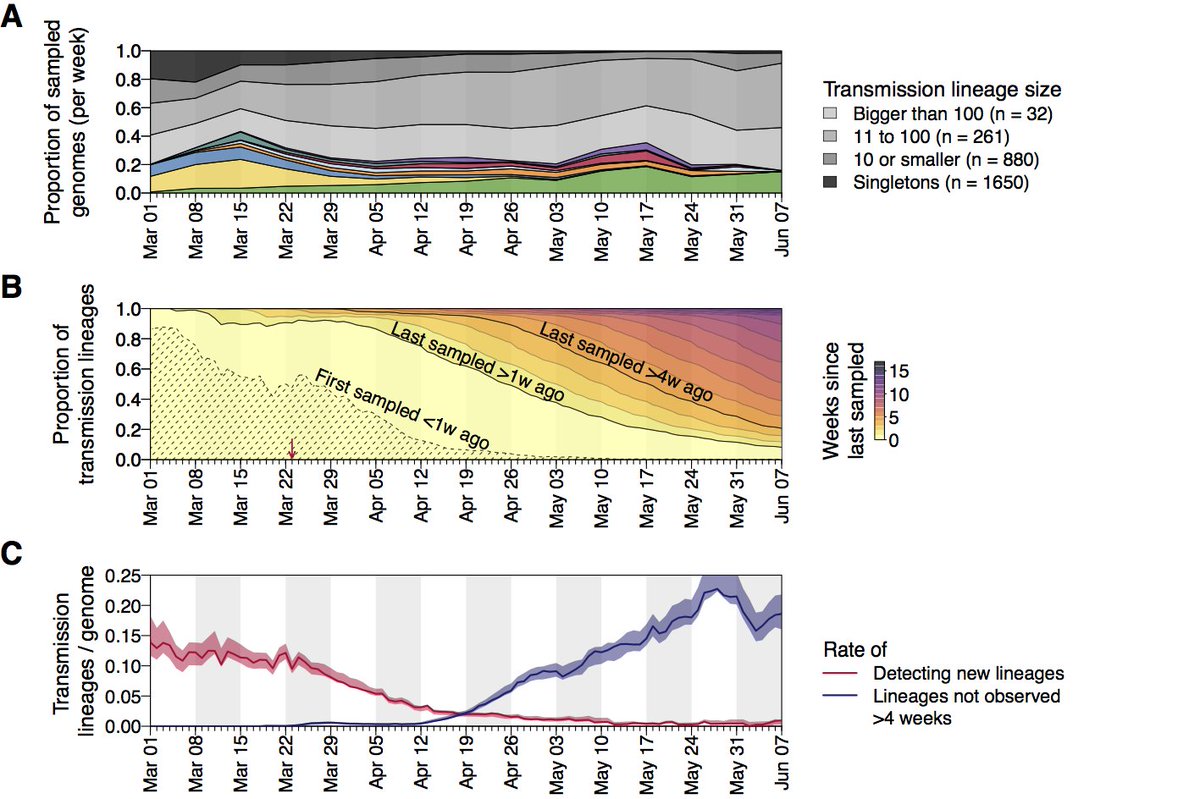
We detect >1000 independently-introduced UK transmission lineages. However we expect this number to be an underestimate. Transmission lineage sizes follow a power-law distribution with the biggest 20% accounting for >75% of all UK genomes in our dataset.
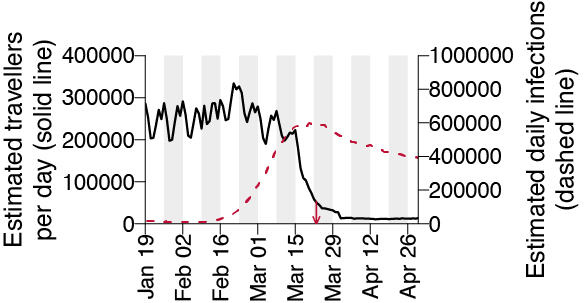
Where do all these lineages come from and what does it mean? Pre-lockdown high travel volumes and few restrictions on arrivals coincided with growing epidemics in Europe, jointly contributing to accelerating the growth of the UK epidemic to exceed contact tracing capacity.
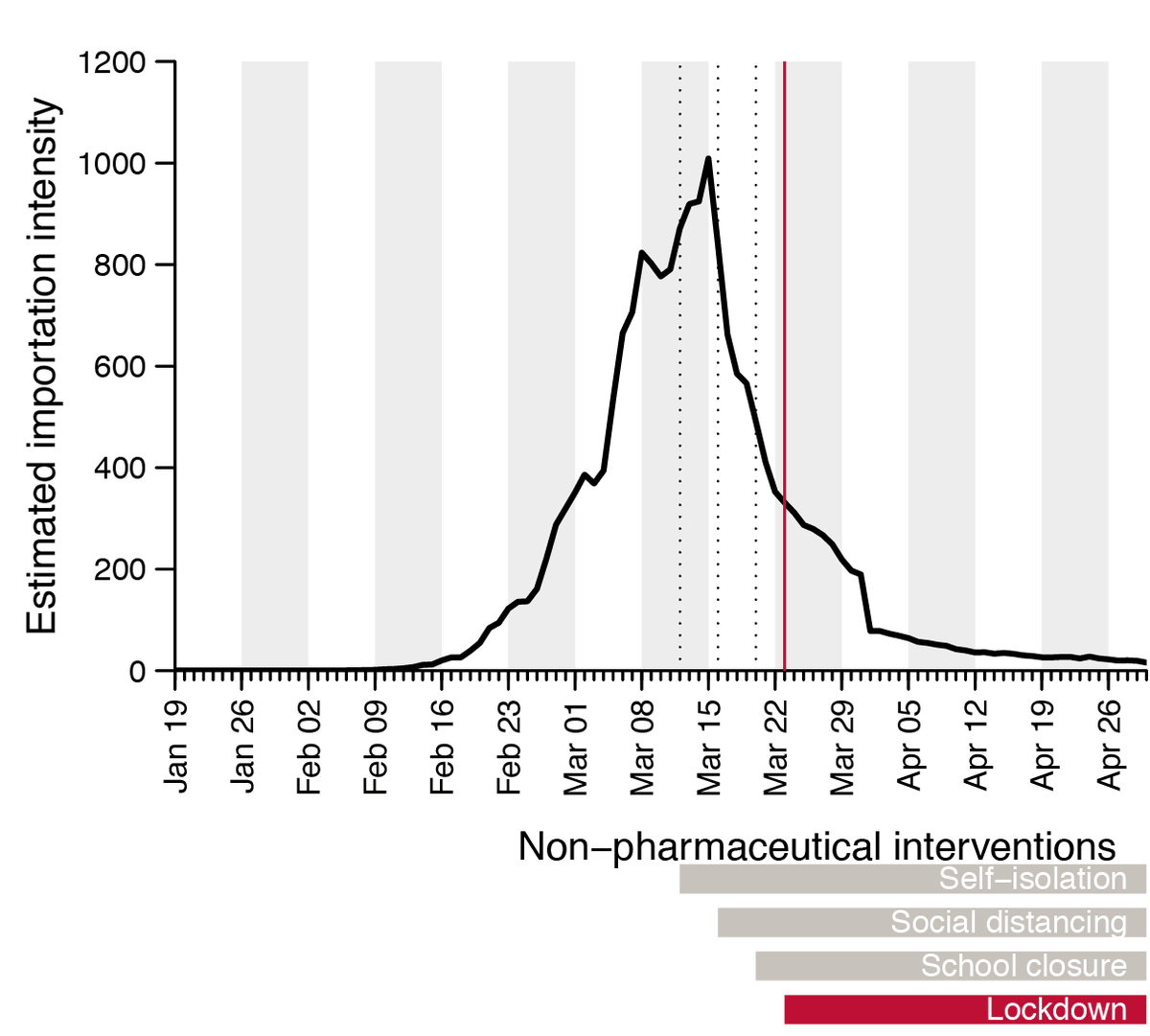
We combined these 2 trends to generate an estimated importation intensity (EII) - an empirical measure of the intensity of #SARSCoV2 importation into the UK - which peaks in mid-March, before the national lockdown.
By combining country-specific EIIs with genomic data and a statistical model we show that the rate and source of introductions changed rapidly through time. In total we find most lineages were imported from Spain, France and Italy, with China only accounting for a tiny minority.
The rate of imports slowed after lockdown, but we find earlier lineages were larger, more dispersed and harder to eliminate, highlighting the importance of rapid or pre-emptive interventions. Overdispersion in transmission likely led to even greater survival of larger lineages.
We expect similar trends occurred in other countries with large epidemics and high travel volumes. These results and methods are changing our understanding of the lineage dynamics of epidemics and are already being used to track the spread of new variants: https://cov-lineages.org/global_report_B.1.1.7.html
Thanks to @EvolveDotZoo, @McCroneIV, @aezarebski, @viralverity, @Chris3Ruis, @arambaut, @MOUGK and many many others for all their hard work on this project!
@B_Gutierrez_G @jnaraghwani @_jordanash @rmcolq @tomrconnor @nmrfaria Ben Jackson @pathogenomenick @AineToole @samstudio8 @krisparag1 @DrEmilyScher @TanyaVasylyeva @erikmvolz @daanensen Alexander Watts @BogochIsaac Kamran Khan
Lastly, this wouldn’t have been possible without the hard work of hundreds of people working for @CovidGenomicsUK, sequencing 1000s of genomes/week. Most are on fixed-term contracts and have been working overtime since March. Really fantastic effort from them!
end;
end;

 Read on Twitter
Read on Twitter