*Functional networks, but what kind? Overlapping and dynamic*
If you're new to neuroscience or to networks, here's a blog post describing how brain networks can be viewed in a more nuanced way. https://cognitionemotion.wordpress.com/2021/01/02/what-kind-of-brain-network-overlapping-and-dynamic/
https://cognitionemotion.wordpress.com/2021/01/02/what-kind-of-brain-network-overlapping-and-dynamic/
If you're new to neuroscience or to networks, here's a blog post describing how brain networks can be viewed in a more nuanced way.
 https://cognitionemotion.wordpress.com/2021/01/02/what-kind-of-brain-network-overlapping-and-dynamic/
https://cognitionemotion.wordpress.com/2021/01/02/what-kind-of-brain-network-overlapping-and-dynamic/
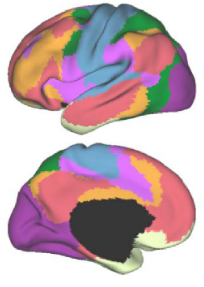
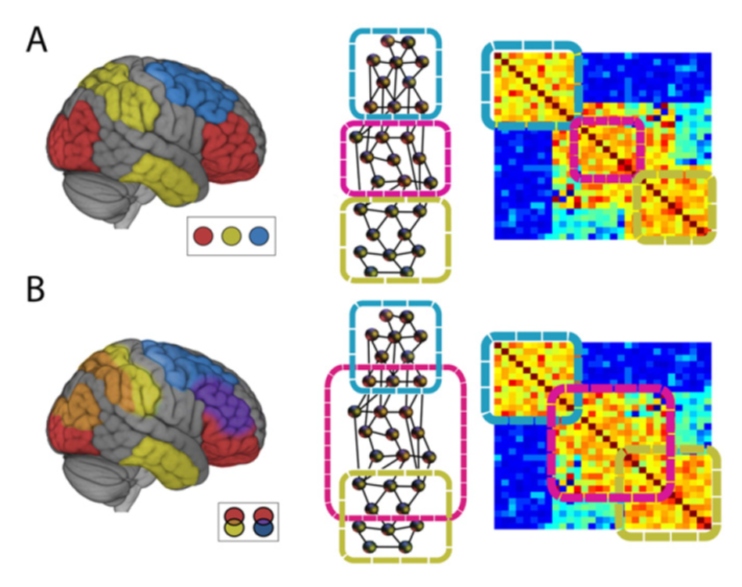
Typically networks in the brain are viewed as disjoint or non-overlapping. This means that a brain region belongs to one and only one network as in the classic Yeo networks.
But we can also partition the brain in *overlapping networks* that allow brain regions to belong to multiple networks simultaneously, so they can dynamically affiliate with different brain regions. (A) Traditional partitioning scheme; (B) overlapping.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4915991/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4915991/
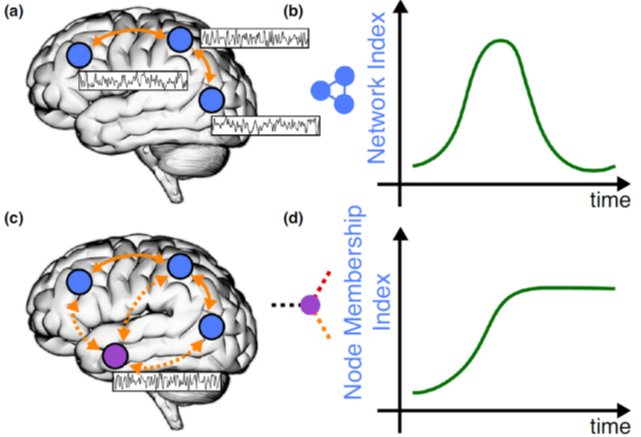
Networks are not static, so any property they have is dynamic and should be thought of in temporal terms. The figure below is from a paper that describes that in more detail:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6003711/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6003711/
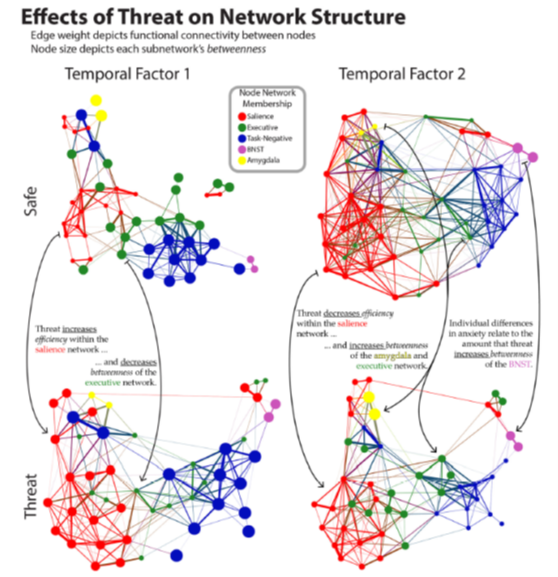
As an example, consider an experiment with temporally extended threat of shock. We found a reorganization of the canonical networks (default, salience, executive) as the "threat block" was experienced.
https://www.jneurosci.org/content/jneuro/34/34/11261.full.pdf
https://www.jneurosci.org/content/jneuro/34/34/11261.full.pdf
So even with fMRI, dynamic properties of networks can be studied if one focuses on temporal scales that are more aligned with the technique (slowly evolving hemodynamic responses).

 Read on Twitter
Read on Twitter