My notes on the #genomics and diagnostics space as we head into 2021. Open to any additional thoughts/ideas.
I believe the move from bulk “grind and find” analysis toward single-cell and single-molecule measurement will be a game changer. $ILMN $PACB $TXG $QTRX
I believe the move from bulk “grind and find” analysis toward single-cell and single-molecule measurement will be a game changer. $ILMN $PACB $TXG $QTRX
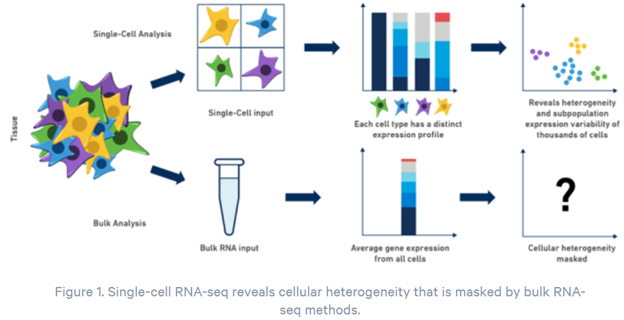
Previously, the only way to measure gene expression was to analyze the information from thousands or millions of cells at a time, resulting in an average measurement across all cells. This hides important cell-to-cell information. https://www.10xgenomics.com/blog/single-cell-rna-seq-an-introductory-overview-and-tools-for-getting-started
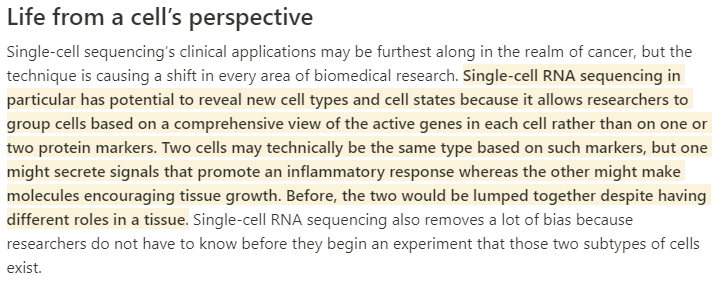
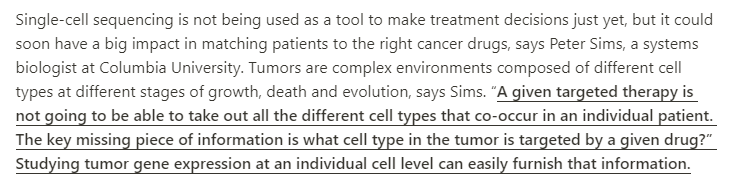
With single-cell sequencing, scientists can now track mutations and changes in gene activity in specific cell types. This article gives you an idea of the potential
"We’re gonna find out about diseases...in a way that we’ve never seen before" https://www.nature.com/articles/d41591-019-00017-6
"We’re gonna find out about diseases...in a way that we’ve never seen before" https://www.nature.com/articles/d41591-019-00017-6
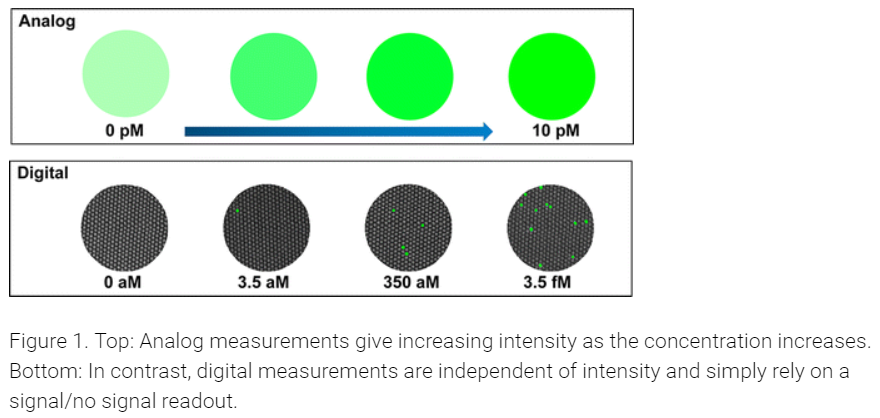
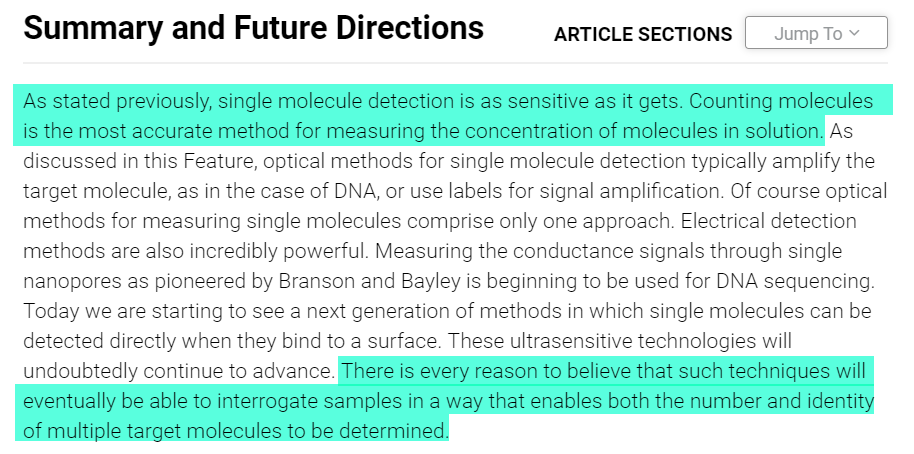
Similarly, measuring individual molecules such as proteins has traditionally relied on analog measurements--the higher the concentration of the molecule being measured, the higher the signal...but the absolute number of molecules present is unknown
Single Molecule Array (SIMOA) technology overcomes this limitation, and is able to provide a digital signal (i.e. a 1 or 0), which can then be counted to provide absolute quantification. This figure highlights the difference between the two
Why does this matter? As explained here:
“the highest resolution measurement one can make is at the single molecule level…in fact, one could argue that the future of all analytical measurements will be molecule counting.”
https://pubs.acs.org/doi/10.1021/ac3027178
“the highest resolution measurement one can make is at the single molecule level…in fact, one could argue that the future of all analytical measurements will be molecule counting.”
https://pubs.acs.org/doi/10.1021/ac3027178

 Read on Twitter
Read on Twitter