Two #SARSCoV2 B.1.1.7 genomes isolated in Canada were just deposited into GISAID today, respectively from BC and Quebec.
Unfortunately, none of the genomes from @BCCDC today was annotated with full sample collection dates. We need this info for molecluar epidemiology. Why?
Unfortunately, none of the genomes from @BCCDC today was annotated with full sample collection dates. We need this info for molecluar epidemiology. Why?
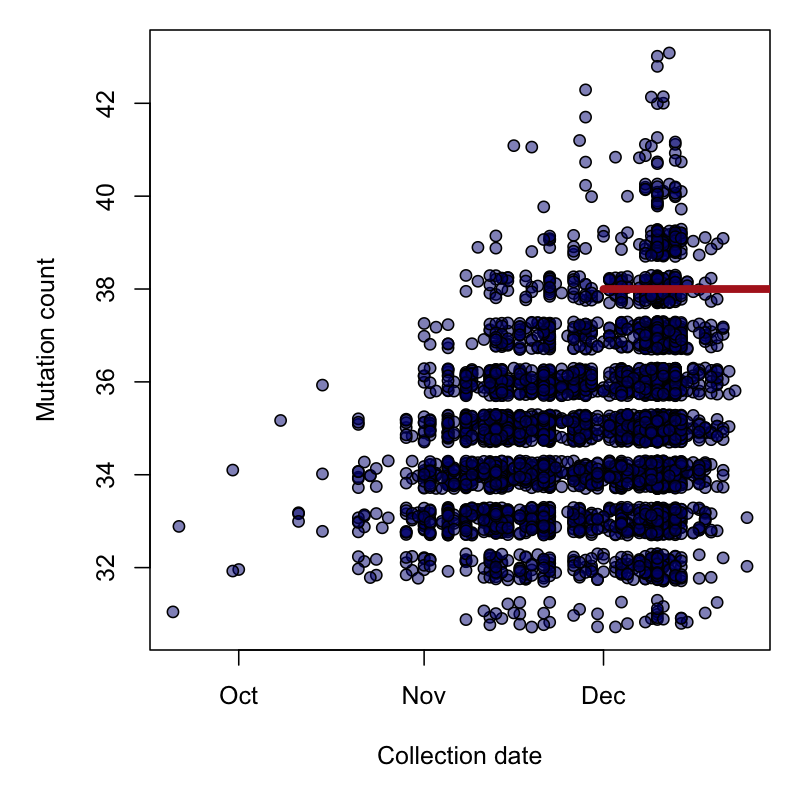
This is a plot of the number of mutations in B.1.1.7 genomes against collection dates for today's database.
I've drawn a red line segment for the BC genome to cover all possible collection dates for this sample. Relative to the time scale for this lineage, this is quite broad.
I've drawn a red line segment for the BC genome to cover all possible collection dates for this sample. Relative to the time scale for this lineage, this is quite broad.
If we can't fix this genome in time, then researchers around the world like @firefoxx66 and @AineToole who are tracking B.1.1.7 can't make use of this info.
When did this lineage enter Canada? How rapidly is it spreading? These are questions we can't answer without dates.
When did this lineage enter Canada? How rapidly is it spreading? These are questions we can't answer without dates.
I can't conceive of a valid reason why we are withholding sample collection dates.
The risk to individual privacy is minimal. Molecular source attribution (reconstructing who infected whom) is nearly impossible for this virus. Benefits to the public are substantial.
So, why?
The risk to individual privacy is minimal. Molecular source attribution (reconstructing who infected whom) is nearly impossible for this virus. Benefits to the public are substantial.
So, why?
(Note, for the above plot I had to exclude genomes with excess missing data, and some with substantially fewer than 30 mutations - possible misclassification by Pangolin @AineToole, @DrEmilyScher?)

 Read on Twitter
Read on Twitter