The latest S:N501 focal build is now up, with data from 28th Dec:
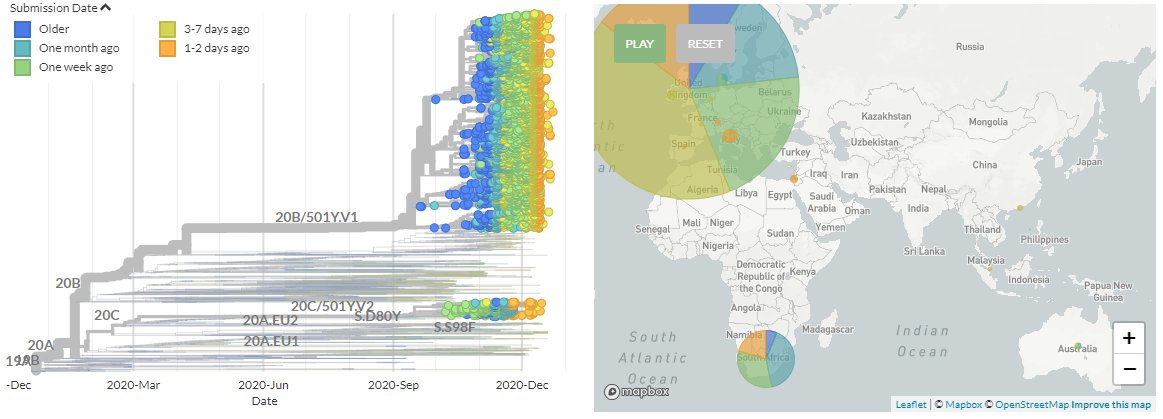
We can see new sequences from Italy, Israel, Spain, France, & Ireland in the SE England variant (501Y.V1) & Switzerland in the South African variant (501Y.V2).
1/8
https://nextstrain.org/groups/neherlab/ncov/S.N501?c=recency&p=grid&r=country
We can see new sequences from Italy, Israel, Spain, France, & Ireland in the SE England variant (501Y.V1) & Switzerland in the South African variant (501Y.V2).
1/8
https://nextstrain.org/groups/neherlab/ncov/S.N501?c=recency&p=grid&r=country
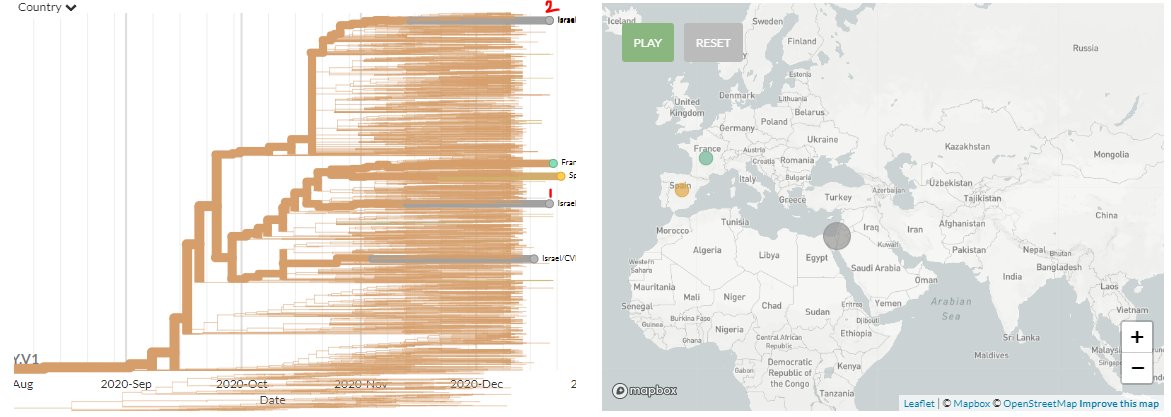
In 501Y.V1 (SE England variant), we can see 1 new sequence from France & Spain, each. The French sample has a travel history to the UK.
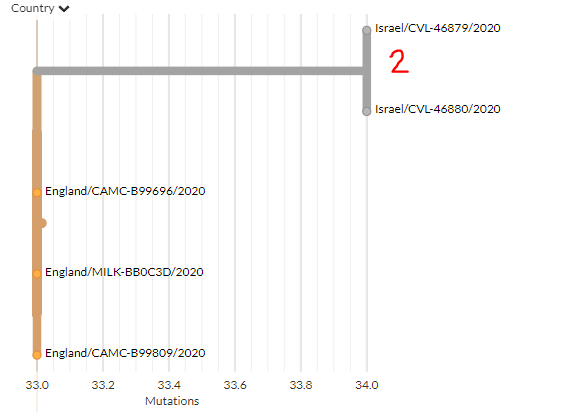
3 new sequences are from Israel (marked 1 & 2). Zooming in on '2' (divergence view) shows they are identical, indicating 1 introduction.
2/8
3 new sequences are from Israel (marked 1 & 2). Zooming in on '2' (divergence view) shows they are identical, indicating 1 introduction.
2/8
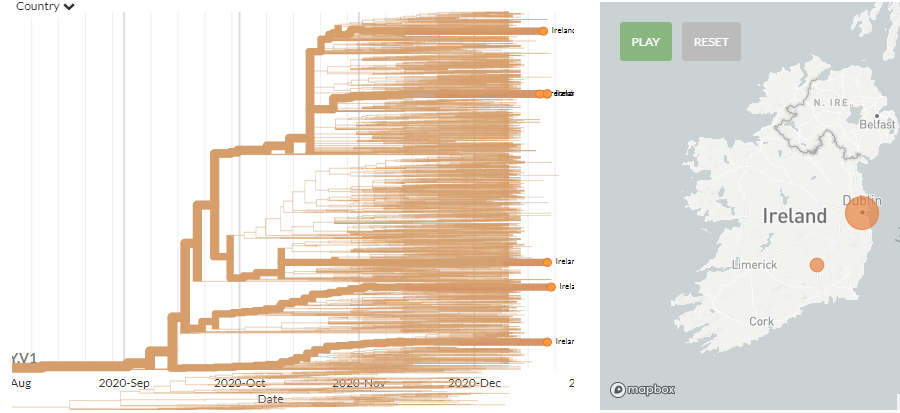
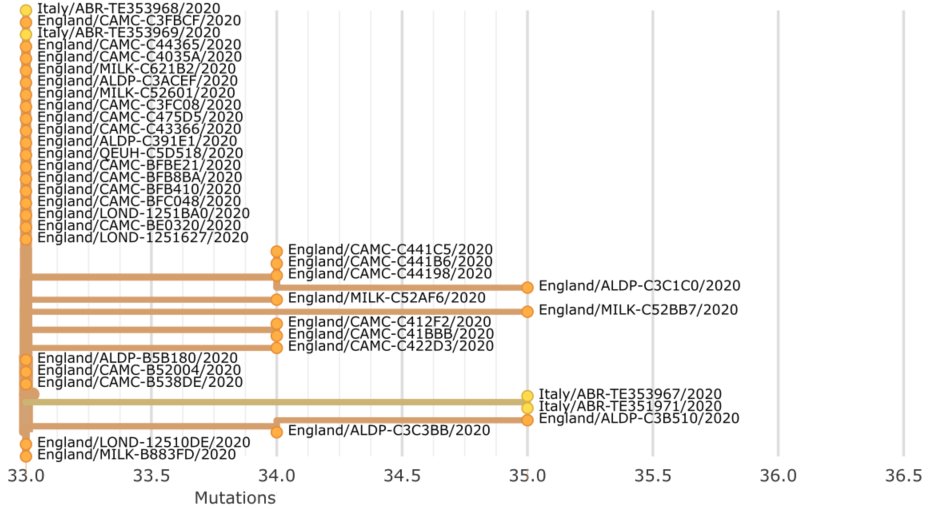
There are 7 new sequences from Ireland, found roughly in 5 places on the tree (4 are singletons, 3 cluster together - 2nd 'group' from the top).
These indicate at least 5 introductions, but we need to zoom in on the 3 that cluster to see more clearly.
3/8
These indicate at least 5 introductions, but we need to zoom in on the 3 that cluster to see more clearly.
3/8
Zooming in to the 3 that cluster together (divergence view), shows it can be difficult to distinguish introductions sometimes, especially as we expect strong travel links between the UK & Ireland, & may only be sampling the total diversity present in Ireland.
4/8
4/8
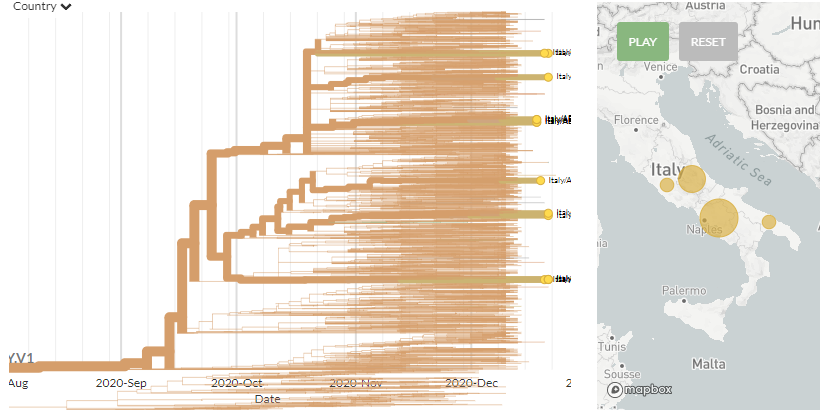
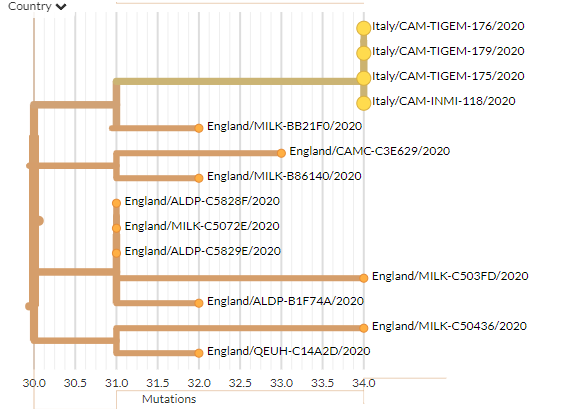
14 new sequences from Italy are scattered across the tree - some alone & some clustering.
Zooming in on a few clusters shows some seem to be linked, while others are harder to determine.
4 seem to be clearly linked as 1 introduction.
5/8
Zooming in on a few clusters shows some seem to be linked, while others are harder to determine.
4 seem to be clearly linked as 1 introduction.
5/8
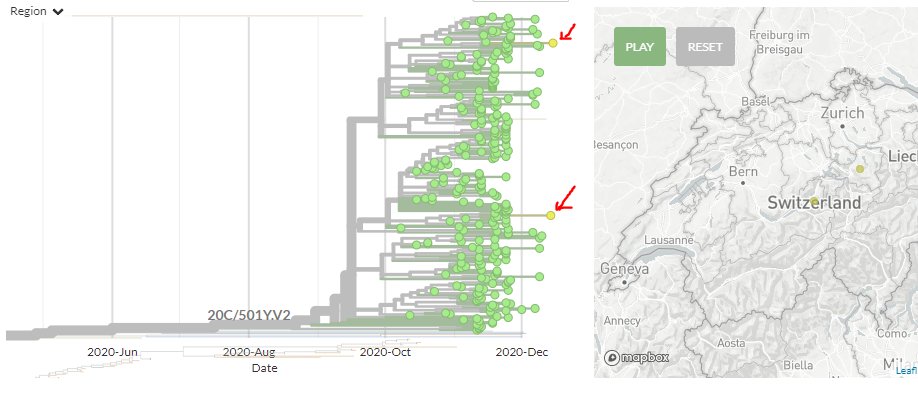
In 501Y.V2 (SA variant), there are 2 new sequences from Switzerland. As they sit quite separately within this cluster, this indicates two separate introductions to Switzerland.
6/8
https://auspice.us/?c=region&f_clade_membership=20B/501Y.V1,20C/501Y.V2&f_country=Switzerland,South%20Africa&p=grid
6/8
https://auspice.us/?c=region&f_clade_membership=20B/501Y.V1,20C/501Y.V2&f_country=Switzerland,South%20Africa&p=grid
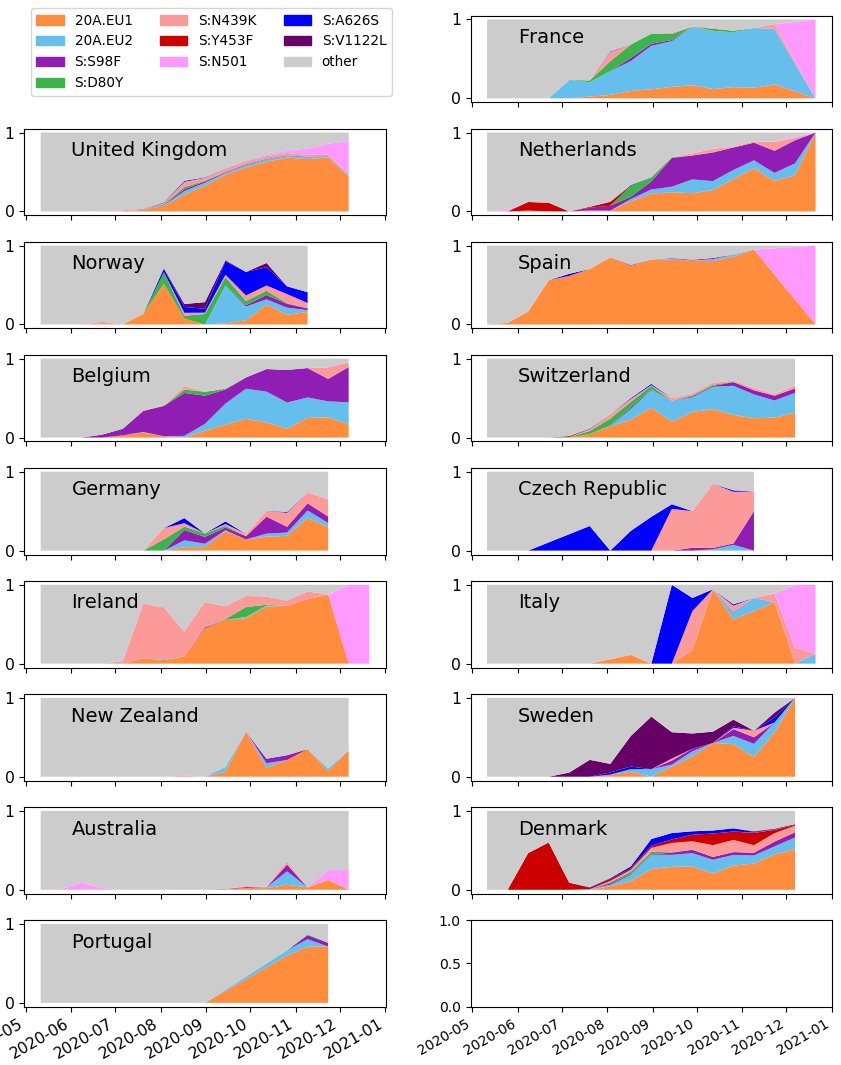
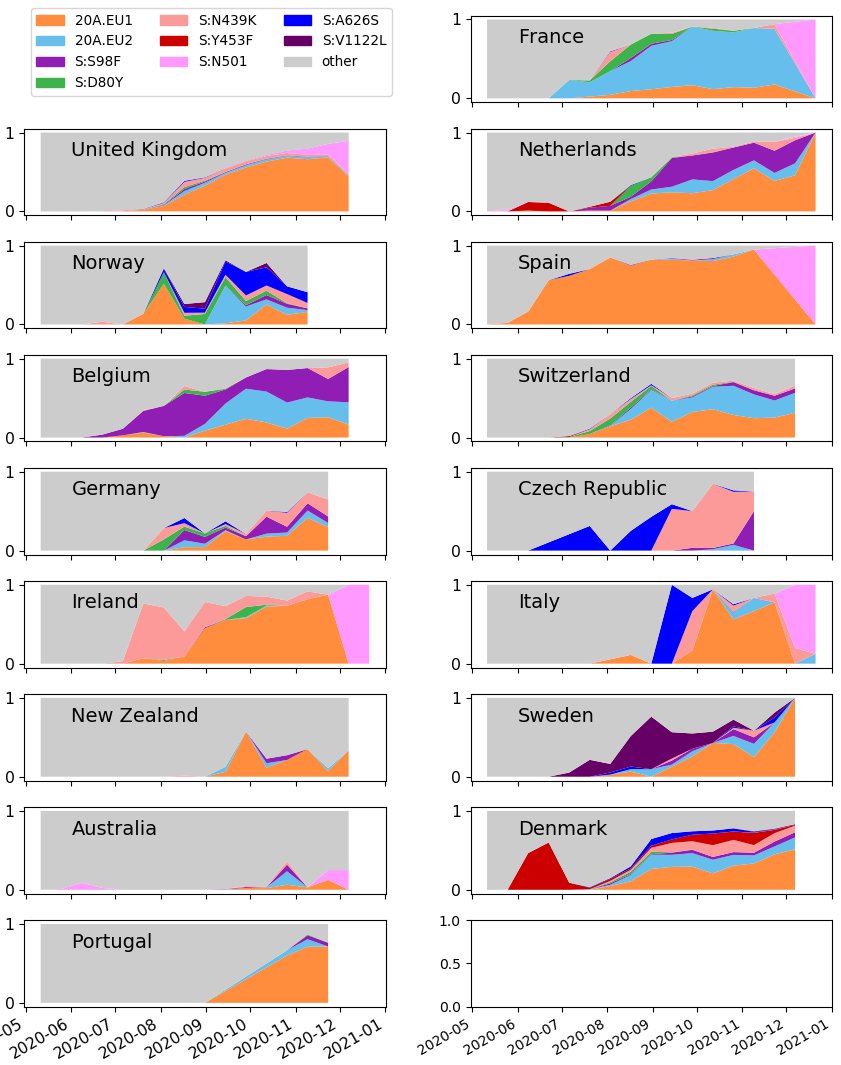
Additionally, a new graph showing the proportion of different variants in selected countries is also up.
Note the sharp in increase of N501 (bright pink) at the end of many graphs: this indicates increased interest in sequencing N501 samples.
7/8
https://github.com/emmahodcroft/covariants/blob/master/country_overview.md
Note the sharp in increase of N501 (bright pink) at the end of many graphs: this indicates increased interest in sequencing N501 samples.
7/8
https://github.com/emmahodcroft/covariants/blob/master/country_overview.md
While more sequencing is great, you can see how switching to preferentially sequencing N501 samples will distort our ability to track other variants, both current & emerging.
Long-term, we need coordinated & regular sequencing of many samples to track #SARSCoV2 variants!
8/8
Long-term, we need coordinated & regular sequencing of many samples to track #SARSCoV2 variants!
8/8

 Read on Twitter
Read on Twitter