SoupX, our method to detect and correct ambient RNA contamination in droplet based single cell sequencing, has been published ( https://academic.oup.com/gigascience/article/9/12/giaa151), code here ( http://github.com/constantAmateur/SoupX/). A thread about the limitations…
Why the limitations? I think it’s reasonably well understood that RNA contamination can be an issue for scRNA-seq (and hopefully that SoupX can help). But I hope understanding the limitations of SoupX will allow more realistic expectations and better use of the tool.
The key point is that removing contaminating counts while leaving true counts alone is a hard problem with no perfect solution. This implies a trade-off: the more aggressively you remove things likely to be contamination, the more real data you’ll remove along the way.
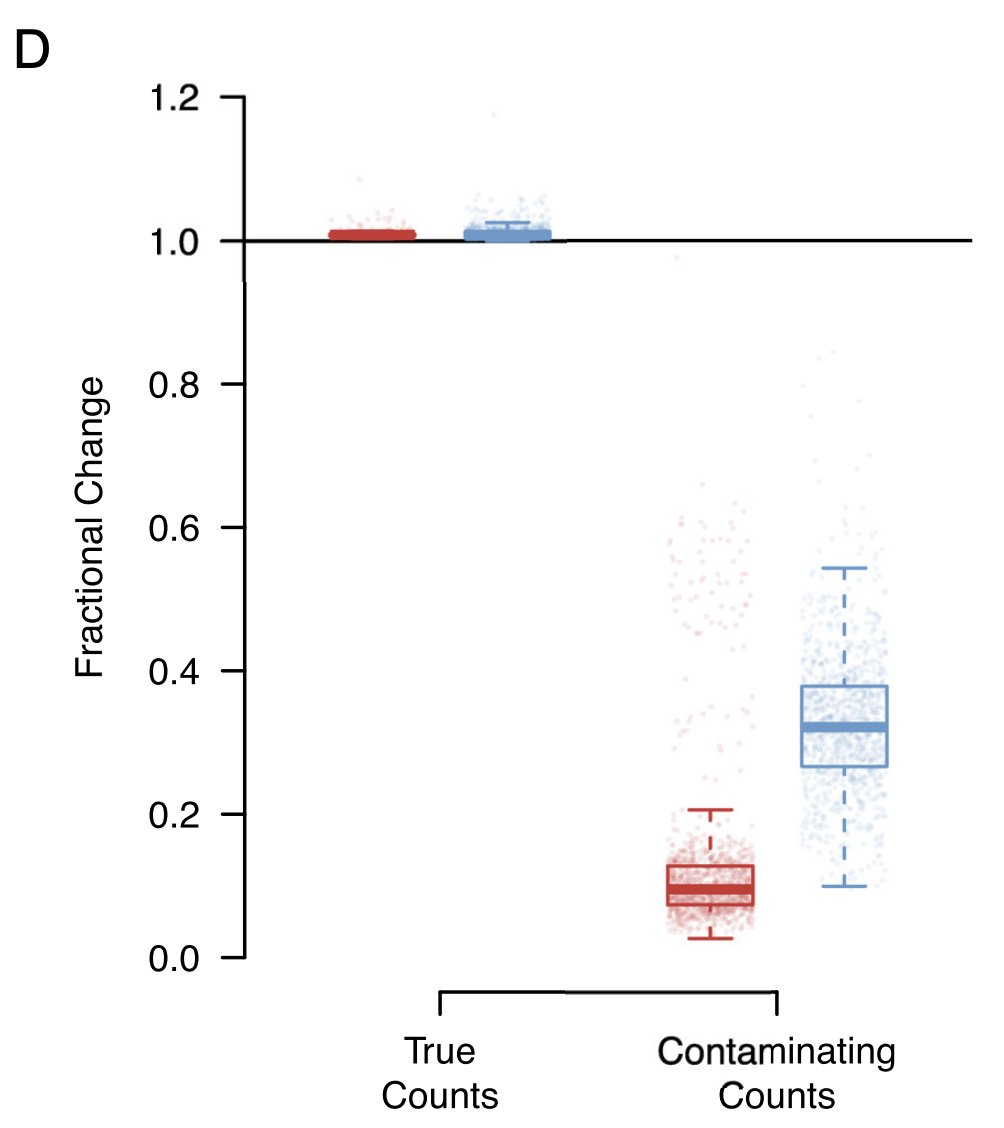
By default, SoupX errs on the side of preserving all true counts even if this means some contamination is left post-correction. This removes around 90% of the contamination and leaves the true counts pretty much untouched in 10X data (red).
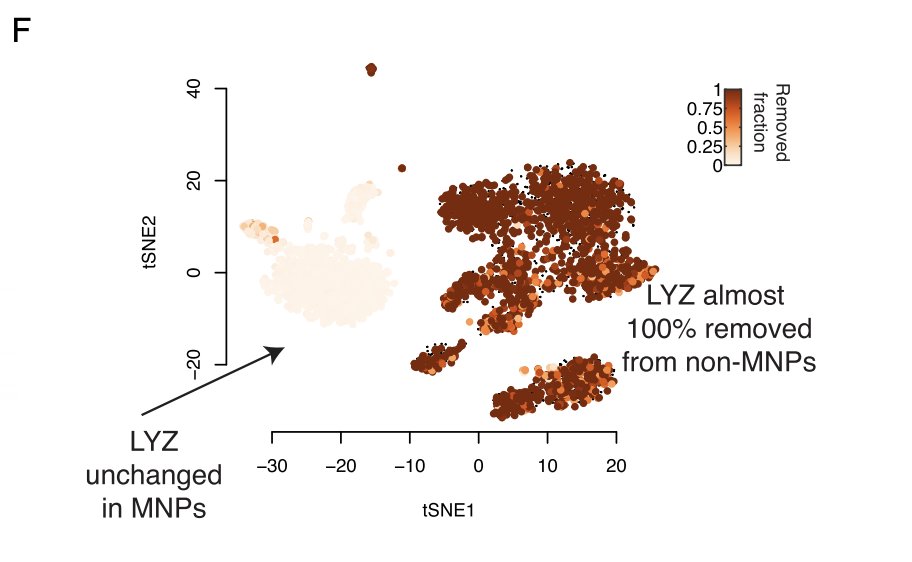
This is often enough to remove all erroneous expression of key genes. But not always, especially for very highly expressed genes like HBB. So it’s entirely possible that you’ll still see some HBB (or whatever your sanity check is) expression in impossible places post correction.
So what should you do if it’s essential that you remove *all* the contamination? You should artificially set the contamination rate to something higher than estimated. This will remove more contamination at the cost of also removing some real expression.
Although throwing out real data sounds bad, it is not as bad as it seems. This is because counts will be removed from things that look like the background. Any gene that is a cell marker will be expressed at a much higher than background and will essentially never be removed.
So as long as you understand the cost of doing it (losing some real expression of non-marker genes), you should feel free to manually set a high contamination rate if your application demands extremely decontaminated data.
For those of you still reading, I wanted to also mention the main innovation in this version: an automated approach to estimating the contamination fraction. This addresses a key complaint about SoupX, that it requires manually specifying gene sets.
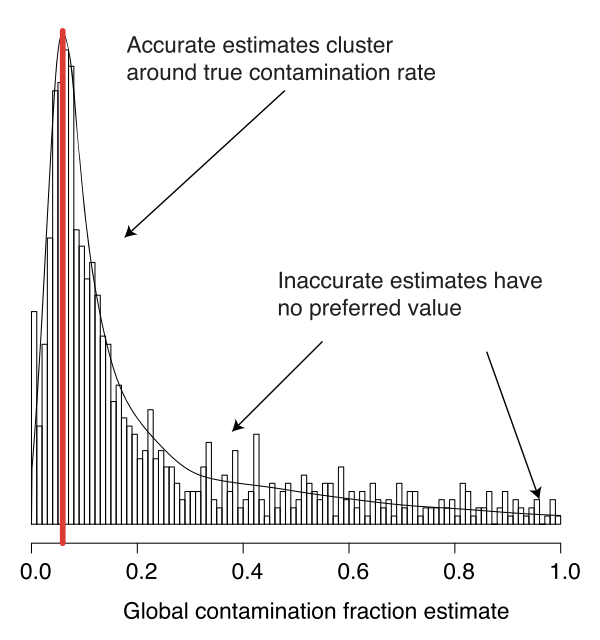
The automated approach uses the observation that marker genes are usually the best genes to estimation contamination and that given many estimates of the contamination, the true estimates should cluster together.
Please use this approach, but recognise that automated doesn’t mean always correct. So if you get a crazily high (or low) estimated contamination rate, treat it with scepticism and consider manually specifying a plausible value or using the old approach.
In particular, this approach is likely to break in very low complexity data (few clusters, low cell #). This is because it relies on multiple distinct clusters being present to identify markers and estimation contamination.
The old approach will probably also struggle with such data. With low complexity data the best approach is probably to manually set the contamination rate to something sensible.
I hope you find SoupX useful. I consider the paper a commitment to support the software, so please raise issues on GitHub. That said, I must prioritise working on new papers on which others careers may depend. But I will get to your question, please be patient and understanding!

 Read on Twitter
Read on Twitter