Happy to share our preprint, by which we propose the novel molecular function of the linker histone H1 in regulation of the mitotic chromosome shape and organization. Here I introduce a historical background for a broad audience. 1/ https://www.biorxiv.org/content/10.1101/2020.12.20.423657v1
First of all, the work was led by a fantastic graduate student, Pavan Choppakatla. He now understands chromosome biology much deeper than I do, and he did write this manuscript! Hi-C was done through a stimulating collaboration with @job_dekker and @DekkerBastiaan. 2/
Recombinant human condensin complexes were provided by Erin Cutts @cutts_dr and Alessandro Vannini. We owed a lot to Erin who patiently shipped her precious reagents twice, as FEDEX ruined her sample in the first round!
HUGE thanks to all collaborators & big congrats Pavan!
3/
HUGE thanks to all collaborators & big congrats Pavan!
3/
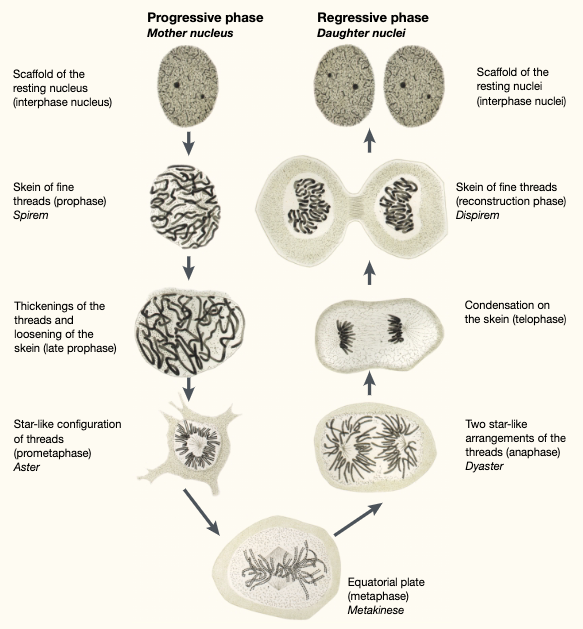
In 1882, Walther Flemming reported that “the scaffold and the network of nucleus transformed in the threads”. Since then, the mechanism behind this morphological transformation of threads, or chromosomes, has been a big question for cell biologists. 4/
https://pubmed.ncbi.nlm.nih.gov/11413469/
https://pubmed.ncbi.nlm.nih.gov/11413469/
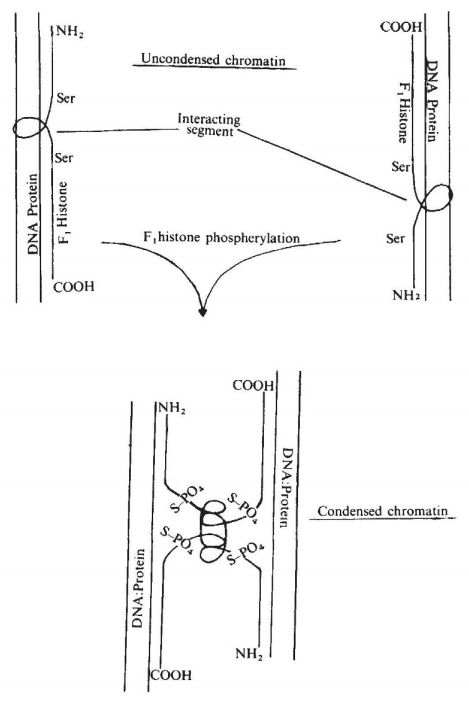
By mid 1970s, it was known that lysine-rich linker histone H1 (F1) is heavily phosphorylated during mitosis. A 1974 Nature paper detected the mitosis-associated histone H1 (F1) kinase activity from the nuclei of the slime mold. 5/ https://pubmed.ncbi.nlm.nih.gov/4361936/
In that 1974 paper, the authors speculated that phosphorylation of H1 would weaken H1-DNA interaction but promote intermolecular interaction between H1 histones to compact chromosomes. However, the molecular function of histone H1 in mitosis remained largely speculative. 6/
In 1988, the hunt for this mitotic H1 kinase was completed by demonstration that the kinases purified from frog & starfish oocytes and HeLa cells are homologous to the fission yeast Cdc2 (aka Cdk1). 7/
https://pubmed.ncbi.nlm.nih.gov/3293803/
https://pubmed.ncbi.nlm.nih.gov/3412486/ https://pubmed.ncbi.nlm.nih.gov/2844417/
https://pubmed.ncbi.nlm.nih.gov/3293803/
https://pubmed.ncbi.nlm.nih.gov/3412486/ https://pubmed.ncbi.nlm.nih.gov/2844417/
The hypothesis that mitotic histone H1 phosphorylation drives mitotic chromosome condensation was once debunked by Keita Ohsumi in 1993 that metaphase chromosome morphology did not appear to be affected by depleting H1 from frog Xenopus egg extracts! 8/ https://pubmed.ncbi.nlm.nih.gov/8266099/
However, in 2005, Tom @MarescaLab and @rebeccaheald showed that depletion of H1 from Xenopus egg extracts actually makes mitotic chromosomes thinner and longer.
But, why thinner and longer?
Our preprint aims to give an answer to this question. 9/
https://pubmed.ncbi.nlm.nih.gov/15967810/
But, why thinner and longer?
Our preprint aims to give an answer to this question. 9/
https://pubmed.ncbi.nlm.nih.gov/15967810/
You now understand how little progress has been made in last 50 years regarding H1 function in mitosis!
Meanwhile condensin complex has been occupying the center stage of mitotic chromosome organization since its discovery by Tatsuya Hirano in 1994.
10/ https://pubmed.ncbi.nlm.nih.gov/7954811/
Meanwhile condensin complex has been occupying the center stage of mitotic chromosome organization since its discovery by Tatsuya Hirano in 1994.
10/ https://pubmed.ncbi.nlm.nih.gov/7954811/
Tatsuya Hirano showed that antibody-mediated inhibition of condensin (or condensin immunodepletion) blocked chromosome thread formation in Xenopus egg extracts.
11/
11/
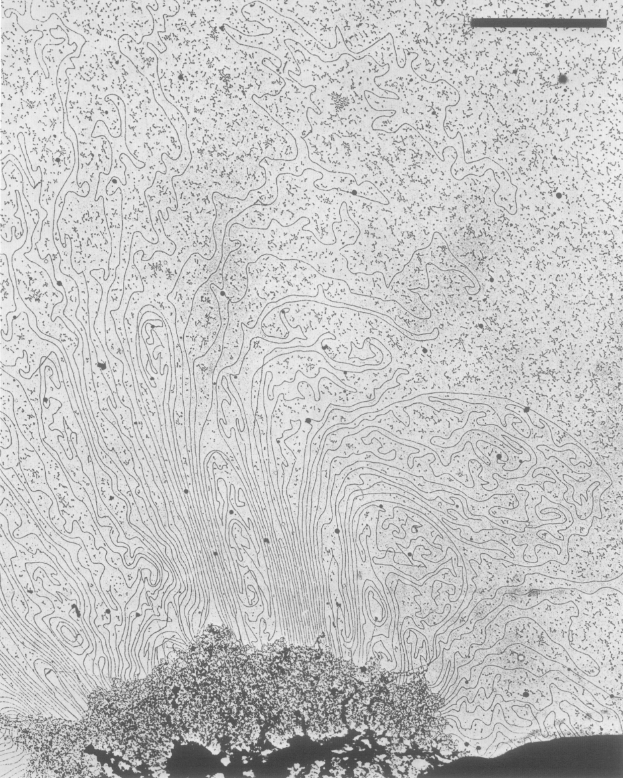
Since Ulrich Laemmli (inventor of SDS-PAGE) discovered removal of histones from chromosomes left a scaffold with 30-90 kb DNA loops in 1977, mitotic chromosomes have been thought to be organized by folding of these long loops. 12/
https://pubmed.ncbi.nlm.nih.gov/922894/
https://pubmed.ncbi.nlm.nih.gov/922894/
Later, Bill Earnshaw showed that the major chromosome “scaffold” proteins are DNA topoisomerase II (topo II) and condensins. Topo II cuts a DNA to pass another DNA strand. Does condensin form a DNA loop, and if so, how?
13/
https://pubmed.ncbi.nlm.nih.gov/2985625 https://pubmed.ncbi.nlm.nih.gov/7929577/
13/
https://pubmed.ncbi.nlm.nih.gov/2985625 https://pubmed.ncbi.nlm.nih.gov/7929577/
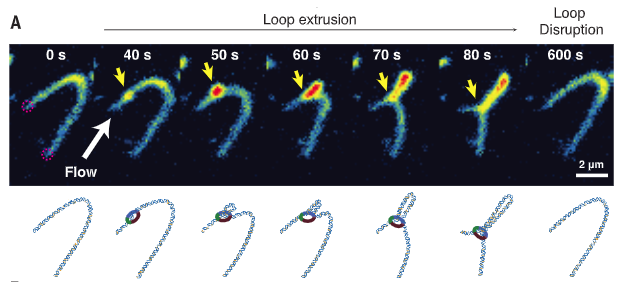
40 years after Laemmli’s observation of DNA loops, labs of @cees_dekker, Christian Hearing, & @DNAcurtain demonstrated that the condensin complex has an ATP-dependent DNA motor and loop exclusion activities!
14/
https://pubmed.ncbi.nlm.nih.gov/28882993/
https://pubmed.ncbi.nlm.nih.gov/29472443/
14/
https://pubmed.ncbi.nlm.nih.gov/28882993/
https://pubmed.ncbi.nlm.nih.gov/29472443/
In 2018, Earnshaw, @leonidmirny & @job_dekker labs published a landmark paper on Hi-C-based analysis of finely synchronized chicken DT40 cells, further supporting the idea that condensins shape mitotic chromosomes via forming layers of DNA loops. 15/
https://pubmed.ncbi.nlm.nih.gov/29348367/
https://pubmed.ncbi.nlm.nih.gov/29348367/
A cool simulation by @golobor, @JohnFMarko & @leonidmirny showing that condensin’s loop exclusion activity & topo II’s strand passing activity are sufficient to drive rod-like chromosome formation. 16/ https://pubmed.ncbi.nlm.nih.gov/27192037/
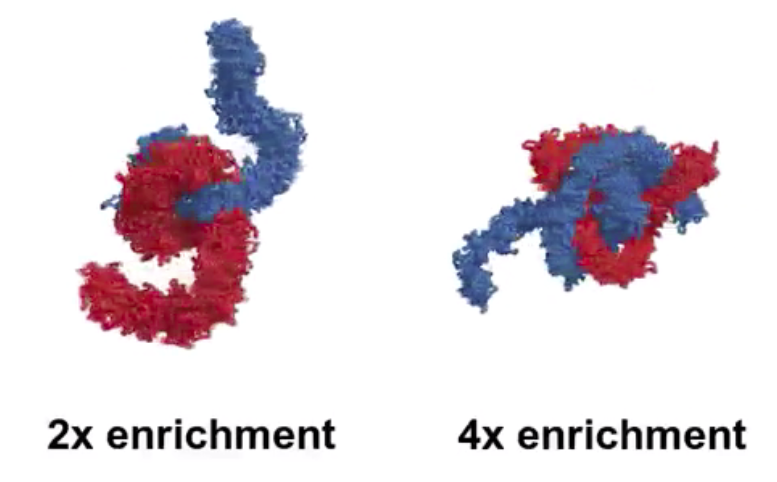
Interestingly, their simulation demonstrated that more the number of condensins on DNA are increased, chromosomes become thinner and longer! 17/
This set up a simple hypothesis;
Did chromosome become thinner and longer in H1-depleted extracts due to higher amount of condensin on chromosomes?
In our preprint, we argue that the answer is YES!
18/
Did chromosome become thinner and longer in H1-depleted extracts due to higher amount of condensin on chromosomes?
In our preprint, we argue that the answer is YES!
18/
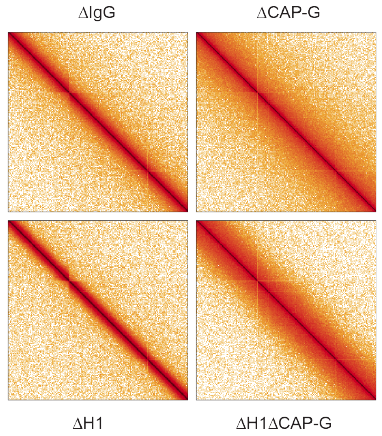
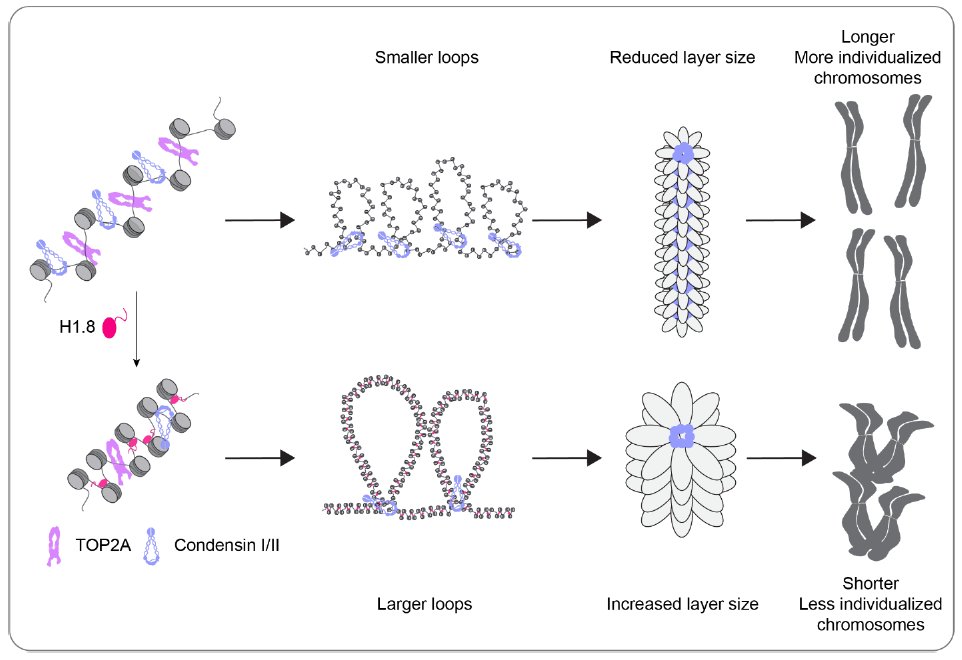
Our Hi-C analysis suggests that DNA loop size indeed becomes shorter upon H1 depletion!
(CAP-G is a subunit of condensin I). 19/
(CAP-G is a subunit of condensin I). 19/
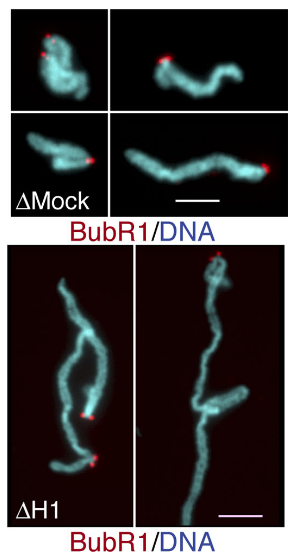
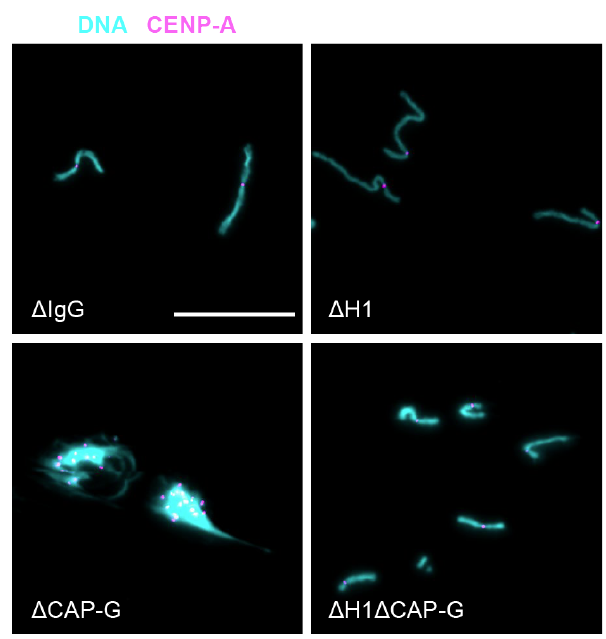
One of the most surprising & exciting moments was when Pavan observed that while condensin I depletion severely inhibited rod-like chromosome formation, co-depletion of H1 rescued this phenotype! We show how come this can be possible! 20/
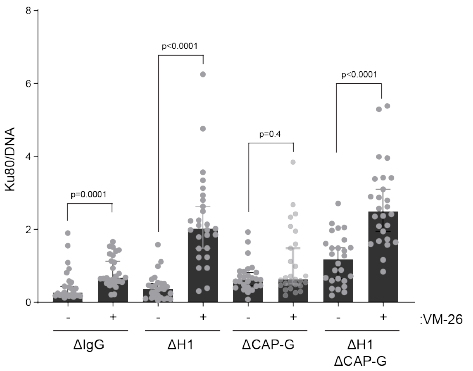
We also found, as has been shown in yeast by Jon Baxter, that condensin is required for mitotic topo II to cut DNA, an activity for chromosome individualization and separation, while now we also show that H1 inhibits topo II activity. 21/
https://pubmed.ncbi.nlm.nih.gov/21393545/
https://pubmed.ncbi.nlm.nih.gov/21393545/
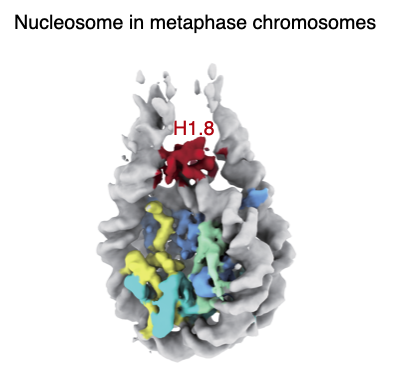
In our recent preprint by @ArmrYshr, we showed that oocyte-specific H1 subtype H1.8 was preferentially loaded on to nucleosome dyad in metaphase chromosomes as compared to interphase chromosomes, but its functional significance was unclear. 22/
https://www.biorxiv.org/content/10.1101/2020.11.12.380386v2
https://www.biorxiv.org/content/10.1101/2020.11.12.380386v2
In our new preprint, we propose that H1.8 suppresses chromatin binding of condensins to keep mitotic chromosome short, and also limits chromosome individualization via suppression of condensins & topo II.
23/
23/
This study opens a number of questions. Can other H1 subtypes also inhibit condensin and topo II? Can H1 inhibit other SMC family complexes, such as cohesin, in interphase? Can chromosome size variations observed during development be explained by differential H1 regulation?
24/
24/
Here comes the end of this thread. It would really great if you could add this preprint in your holiday reading list. Please give us any feedbacks! Happy holidays!
25/25
25/25

 Read on Twitter
Read on Twitter