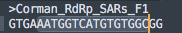
I've been asked this many times. So tweet thread it is.
Do the Drosten primers hit the human genome?
Why do some people claim they hit 100%?
TL/DR: Here is the output from BLAST
https://mega.nz/file/tBpQjLrY#9STxgLOiCc9SJ3FdB1C9zo2UvBbt83hlLyTKLG3Ohz0
Here are the alignments from BLAST
https://mega.nz/file/0A5QgTBI#1MnMxaqM5q9-QwWk-0XrOwVLcorGLQymwbU6aUYFezg
Do the Drosten primers hit the human genome?
Why do some people claim they hit 100%?
TL/DR: Here is the output from BLAST
https://mega.nz/file/tBpQjLrY#9STxgLOiCc9SJ3FdB1C9zo2UvBbt83hlLyTKLG3Ohz0
Here are the alignments from BLAST
https://mega.nz/file/0A5QgTBI#1MnMxaqM5q9-QwWk-0XrOwVLcorGLQymwbU6aUYFezg
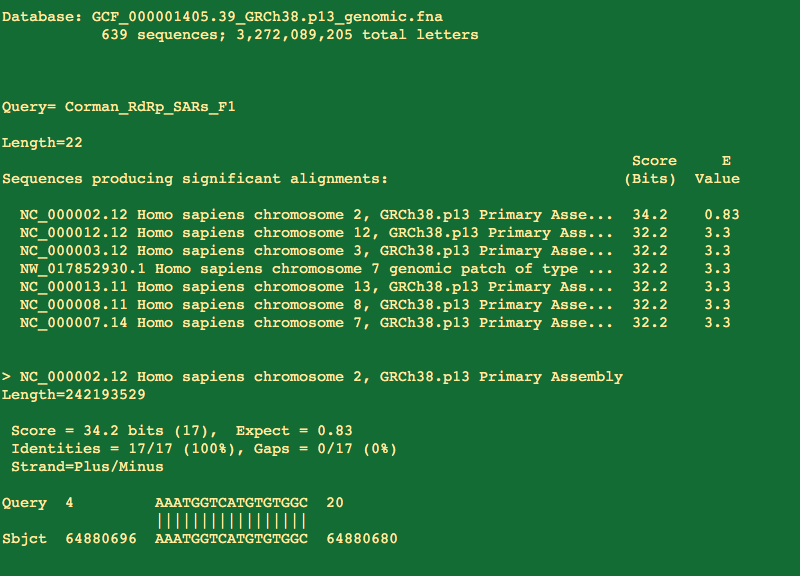
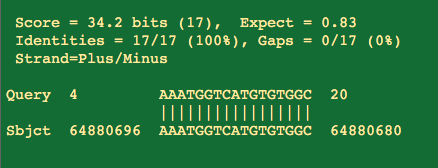
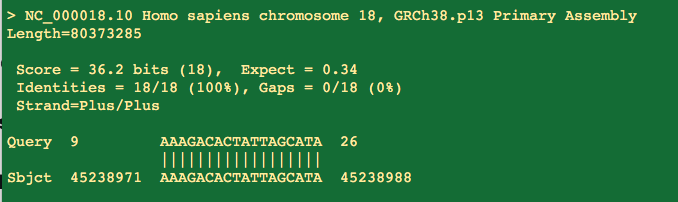
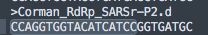
So the first thing people get excited about is that portions of the primers will hit with 100%with 17/17 residues hitting the human genome.
But you will notice the 3' end of the primer doesn't match and that is the business end of the primer.
But you will notice the 3' end of the primer doesn't match and that is the business end of the primer.
That 17 bases represents the middle of the primer highlighted below and doesn't include the 3 prime end. Not ideal but not lethal.
In addition to this primer landing, PCR also requires the Reverse primer land with high specificity on its 3' end.
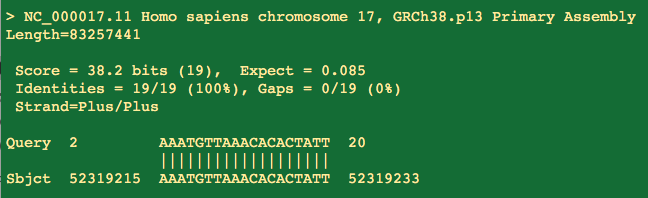
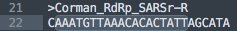
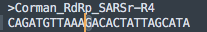
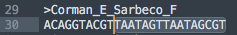
What do we have for the Reverse primer?
Looks like a 100% hit! Controversy! Conspiracy!
Not quite. This is another primer that doesnt land on the 3 prime end.
What do we have for the Reverse primer?
Looks like a 100% hit! Controversy! Conspiracy!
Not quite. This is another primer that doesnt land on the 3 prime end.
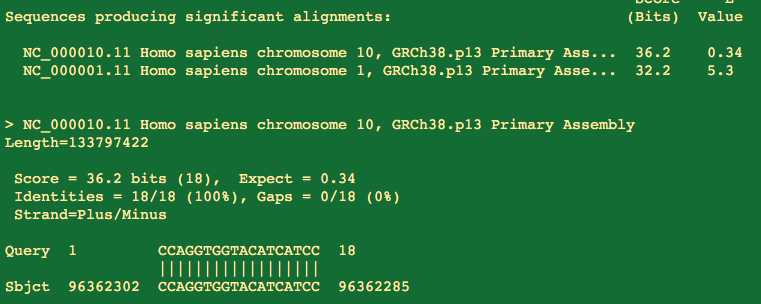
Alas, 6bp on the 3 prime end don't match. Polymerases wont extend this.
Also note its on a different chromosome than the forward hit so even if they landed perfectly on their 3 prime ends, exponential amplification is not possible.
Also note its on a different chromosome than the forward hit so even if they landed perfectly on their 3 prime ends, exponential amplification is not possible.
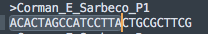
This where the degenerate primers (wobble bases) from Drosten get him into trouble. This is also the Reverse primer but there are more than 1 version of it. There is a C->G variant in the primer that then makes a better match for the primer on Chr. 18 with a 3 prime match.
Just to be thorough, Lets look at the probes. I say Probes because the RdRp probe is a nightmare with multiple degenerate bases and hairpins. The 5 prime end of the probe does anneal. This end of the molecule has the FAM dye on it and will be liberated even if partially annealed
The polymerase extending from a primer would run into the 5' of the probe 1st and hydrolyze the FAM dye from the quencher on the 3' end of the probe. But the homology is not even on the same chromosome as the Forward and Reverse primers. Not ideal but unlikely to pose problems.
Onto the E gene assay.
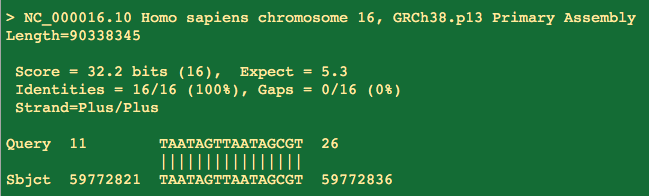
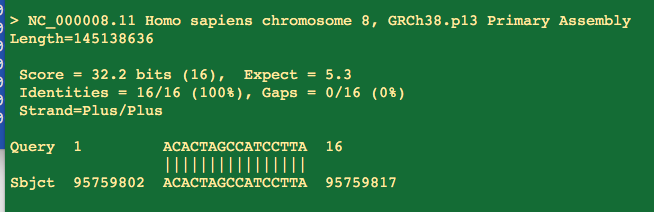
It does have a 16 bp perfect homology to Human chromosome 16. 3 prime end matches.
Does the reverse land in the same neighborhood?
It does have a 16 bp perfect homology to Human chromosome 16. 3 prime end matches.
Does the reverse land in the same neighborhood?
16 bases is a bit short but we must keep in mind there is a 10 minute 55C RT step prior to PCR which is lower than the melting temperature of the PCR reaction. This is where the most off target noise is generated.
There is a place on Chrom. 8 for the E gene probe to land for 16 bases on its 5 prime end but again... This isnt in the right neighborhood to create exponential PCR with signal.
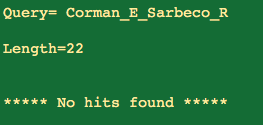
The E gene Reverse is a big goose egg.
So, these homologies are not ideal but are unlikely to be the source of the False positives reported by the test.
BUT.. when your primers can land on human DNA they can extend and create very diverse 3 prime ends of your primer pool.
So, these homologies are not ideal but are unlikely to be the source of the False positives reported by the test.
BUT.. when your primers can land on human DNA they can extend and create very diverse 3 prime ends of your primer pool.
This primer promiscuity can create other unexpected results. Its better to screen your primer against these known artifacts to prevent your primers from landing off target and then extending. While the extension may not amplify your target, it may change the ends of your primers.
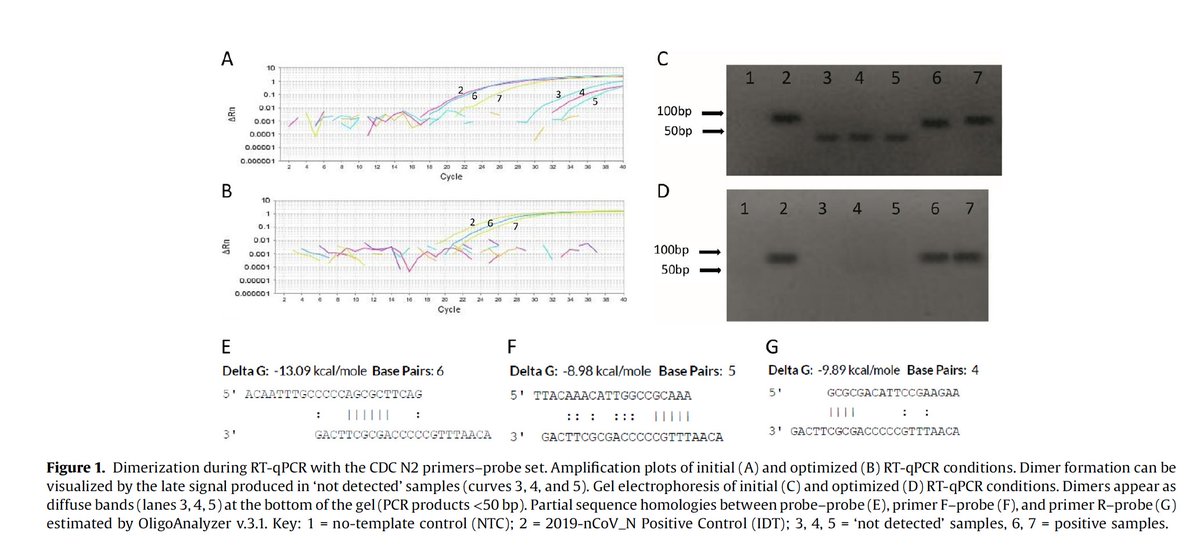
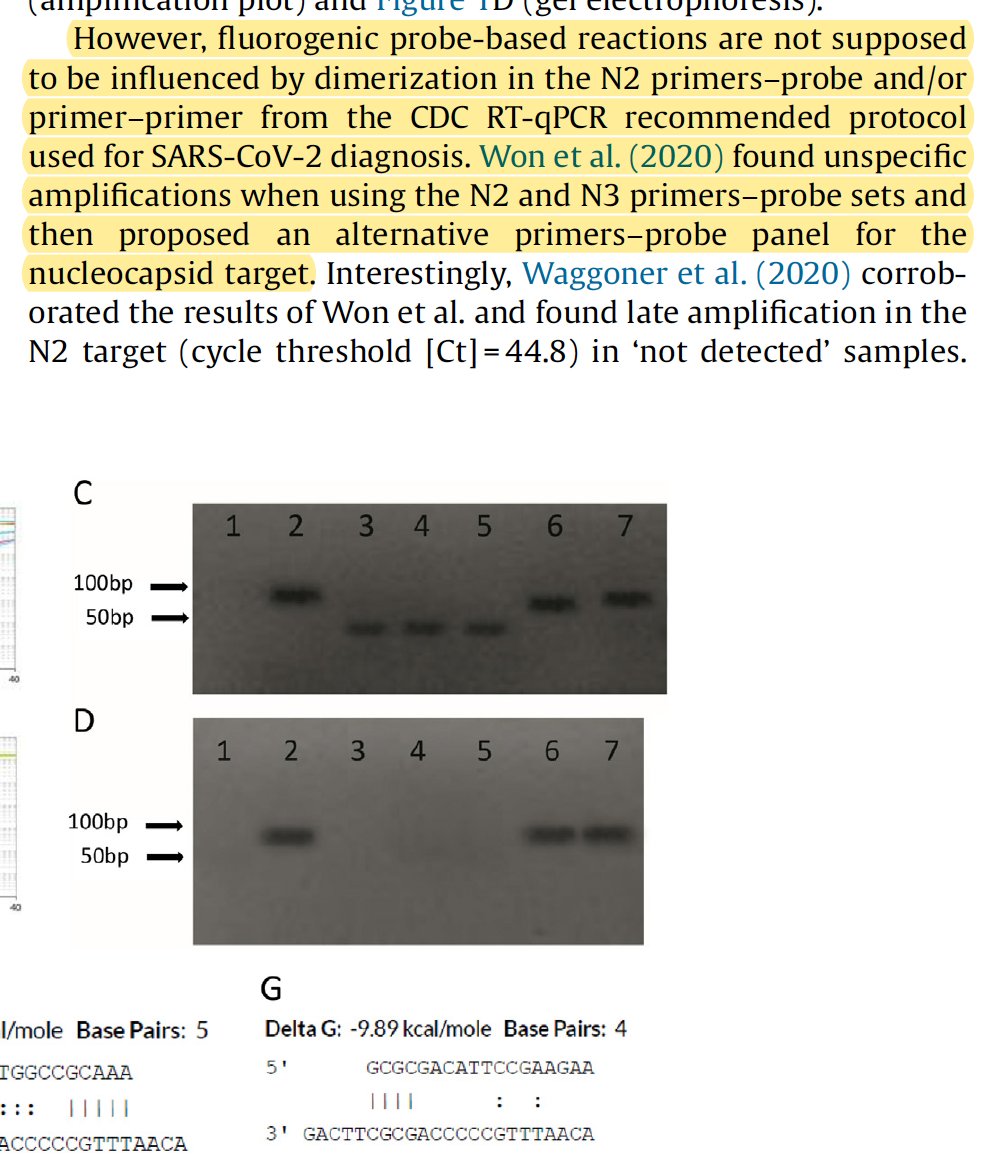
If there is one take away from this thread, the human homologies are not lethal. They are a sign of sloppy primer design but when you have water samples lighting up (no human DNA), you have to look at primer dimers.
When PCR specialists tell you Taqman or probe based qPCR cant..
When PCR specialists tell you Taqman or probe based qPCR cant..
Cant make signal in qPCR, they simply don't have a lot of experience making many assays and haven't seen it yet. As this paper from Jaeger clearly shows.. This can indeed happen and its fare more likely to happen if your primers bind to themselves. https://www.sciencedirect.com/science/article/pii/S1201971220322839

 Read on Twitter
Read on Twitter