You’re the medical registrar on-call and receive a call from the psychiatry SHO as a patient has an adjusted calcium of 1.76.
This is a 31 year old male currently admitted with major depressive episode. He has reported parasthesiae and muscle cramps prompting the blood tests.
I’m sure you all know how to treat acute hypocalcaemia so that’s not the focus of this thread.
His medical history is remarkable for anxiety and depression as well as ADHD.
His medical history is remarkable for anxiety and depression as well as ADHD.
Most recently, he has become a father but this has led to a worsening of his anxiety as his son is due to undergo a major heart operation.
On examination, you notice some distinctive facial features: slightly low set ears, thin upper lip and, strangely, a bifid uvula (doesn’t everybody check the uvula?).
Which of the following investigations is most likely to identify the genetic abnormality:
What is most likely the underlying cause for his hypocalcaemia?
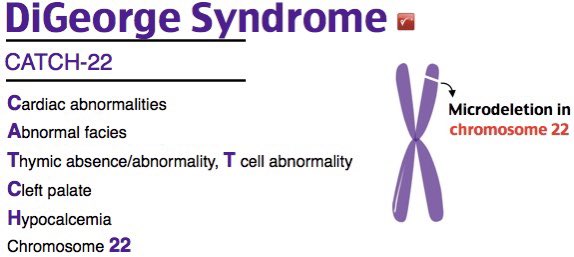
This is DiGeorge Syndrome, also known as 22q11.2 deletion Syndrome (that’s twenty two ‘q’ one one, NOT ... ‘q’ eleven - region 1, band 1, sub-band 2 of the long arm of chromosome 22). This is a deletion usually of around 3Mb (million base pairs).
A good mnemonic is CATCH22 for clinical features but be mindful not every patient will have all these features.
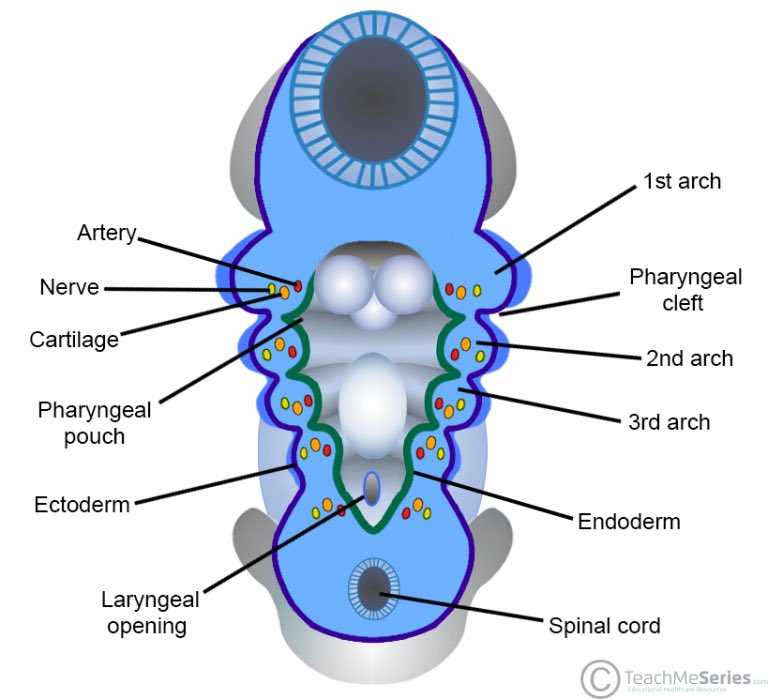
This disorder affects development of the 3rd and 4th branchial arches, which give rise to the parathyroid glands and thymus.
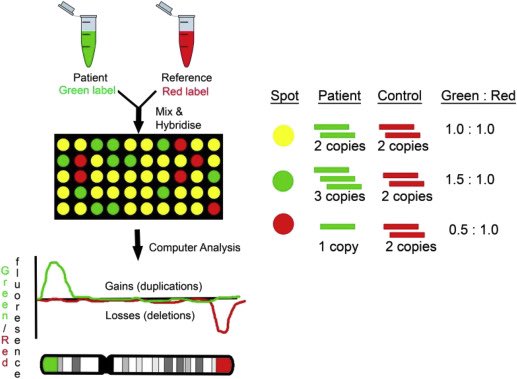
A microarray is a good way to pick this up and will pick up any other gains or losses over around 120Kb (thousand bases). A karyotype won’t pick it up as the resolution is limited to about 5Mb. FISH would be most specific.
A microarray uses thousands (sometimes millions) of probes on a glass slide that are specific for regions of the genome. Fluorescently-labelled DNA is mixed with control, fragmented and hybridised with the slide. Losses and gains are detected through signal intensity.
You could pick this up with whole genome sequencing but that’s expensive and also technically / bioinformatically tricky.
22q11.2 deletion Syndrome affects 1 in 4000 births and is a good example of variable expressivity - people, even in the same family, with this deletion can manifest different problems.
In this case, the patient’s son has inherited the deletion which has caused a congenital heart defect (most likely conotruncal) whilst the patient has ADHD, hypocalcaemia and a bifid uvula.
This is different to penetrance which is the measurement of the proportion of people with a genetic variant that express the disease phenotype.
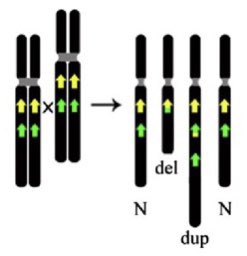
The 3Mb deletion is recurrent (occurs in unrelated individuals) because it is flanked by regions of repetitive DNA which cause misalignment during meiosis resulting in unequal crossing over. The technical term is Non-allelic Homologous Recombination (NAHR).
NAHR causes unequal exchange of material between paired chromosomes so one will gain (duplication) and one will lose (deletion) material. It’s not surprising then that there is a sister duplication disorder to 22q11.2 deletion syndrome.
You’re probably right in thinking ‘I’ll never see a delayed diagnosis DiGeorge presenting as acute hypocalcaemia’ but it has happened and it’s a genomic disorder you are quite likely to see.
Bonus round:
This patient’s son is found to have thrombocytopenia. Which of the following might you suspect?
This patient’s son is found to have thrombocytopenia. Which of the following might you suspect?
That’s right! The GPIbβ gene is located within the DiGeorge critical region and hence deleted. A second mutation on the maternal non-deleted allele has resulted in Bernard-Soulier syndrome - he is ‘hemizygous.’
So that’s 22q11.2 deletion syndrome. It’s important to remember many of the technologies we have today are new and there may be adult patients who were not investigated as children. They are also at risk of developing psychiatric disorders.

 Read on Twitter
Read on Twitter