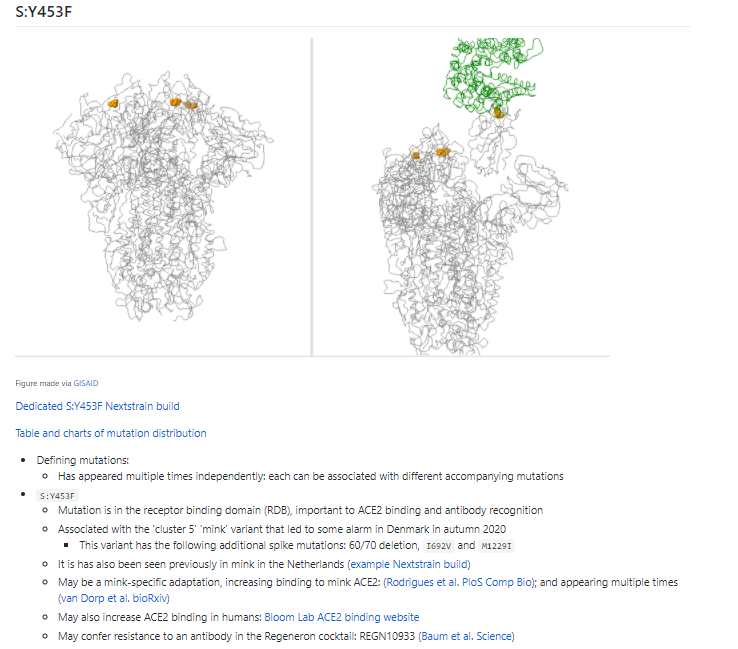
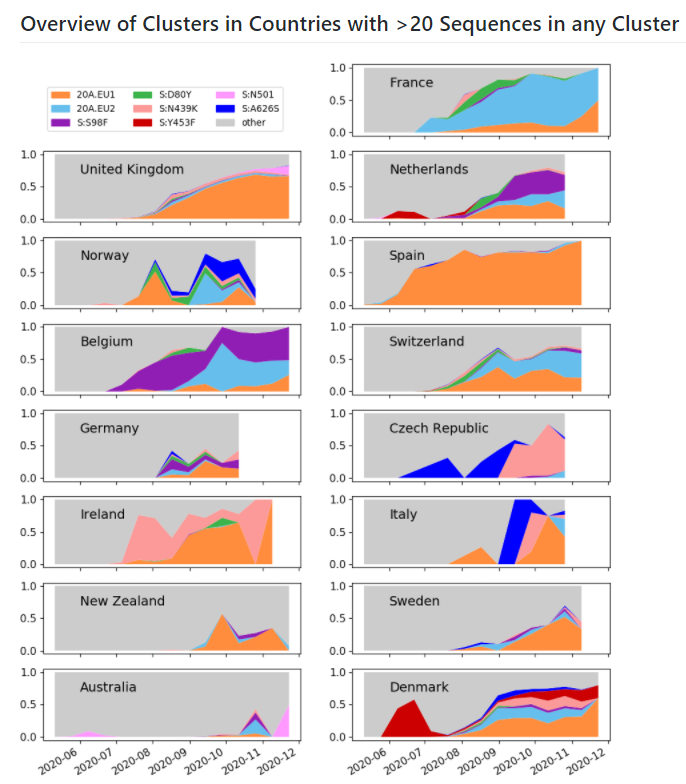
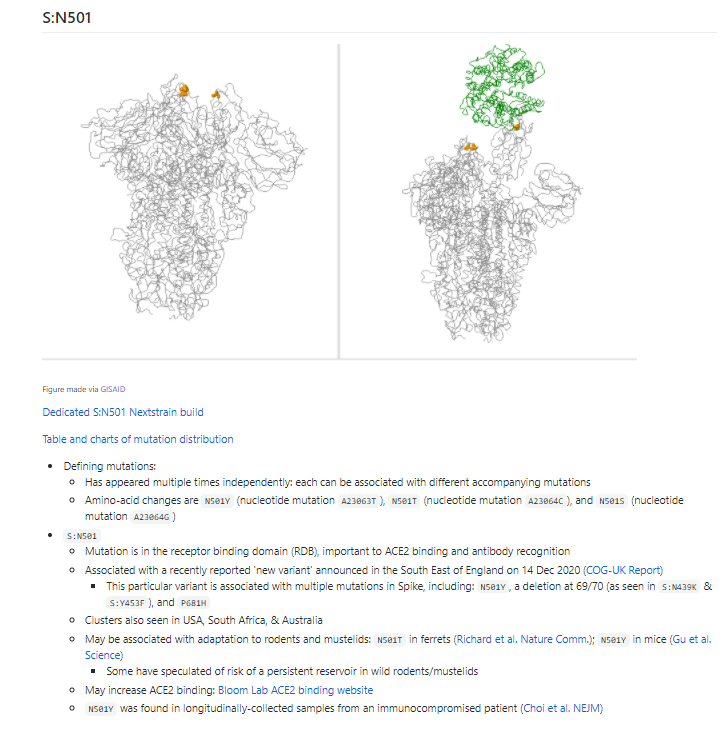
There are many, many #SARSCoV2 #COVID19 variants out there! A few have caught the attention of scientists in Europe - we're trying to monitor them!
I'll be maintaining updated information here:
https://github.com/emmahodcroft/cluster_scripts/blob/master/README.md
#newvariant
1/7
I'll be maintaining updated information here:
https://github.com/emmahodcroft/cluster_scripts/blob/master/README.md
#newvariant
1/7
This site includes a bit about what we know about specific mutations/clusters, including counts of sequences, some graphs, and links to dedicated @nextstrain runs (maintained by @richardneher Lab & myself) focused on each mutation
2/7
2/7
You can also check out graphs of the prevalence of the clusters in various countries:
3/7
https://github.com/emmahodcroft/cluster_scripts/blob/master/country_overview.md
3/7
https://github.com/emmahodcroft/cluster_scripts/blob/master/country_overview.md
The focused @nextstrain builds pull out all sequences with the relevant mutation/part of the relevant cluster & then add closely related background. So they allow a fantastic way to take a close-up look at the variants.
Each is linked in the mutation's 'section'!
4/7
Each is linked in the mutation's 'section'!
4/7
I'll be doing my best to keep information updated, but I *welcome* PRs & links to include the latest publications, preprints, & research!
Our best work on #SARCoV2 #COVID19 is done collaboratively! No one person can keep up with it all
I aim to update at least weekly.
5/7
Our best work on #SARCoV2 #COVID19 is done collaboratively! No one person can keep up with it all

I aim to update at least weekly.
5/7
This page is currently incomplete: I was working on this already - but I've rushed this through to get it good enough to release, as I hope it will be useful straight away - but please be patient as I continue to add information!
6/7
6/7
And yes, there's a section on S:N501: 
I'll be tweeting some of the information here on my original thread on N501Y shortly!
7/7
#newvariant
https://github.com/emmahodcroft/cluster_scripts/blob/master/README.md#sn501

I'll be tweeting some of the information here on my original thread on N501Y shortly!
7/7
#newvariant
https://github.com/emmahodcroft/cluster_scripts/blob/master/README.md#sn501

 Read on Twitter
Read on Twitter