With all respect for the authors, I think this is an extremely poor paper. The good thing is, I also think it's actually very easy to disprove. A few thoughts bellow https://www.biorxiv.org/content/10.1101/2020.12.12.422516v1
I will not comment on the cell cultures overexpression experiments with LINE or HIV-RT (Fig 2) because they are meaningless , and Figure 3 is mainly unrelated correlation
But let's say a word about the very weak Figure 1. They say they can find chimeric RNA-seq reads between SARS-2 and the human genome. And then make the extraordinary claim that these reads originate from SARS-2 sequences that were reverse-transcribed and integrated in the genome
This is a strong, dangerous, and largely unsupported claim, especially since there is an alternative hypothesis that is in my opinion way more likely. I would bet a lot of money that these read originate from RNA recombination between SARS-2 and human mRNAs.
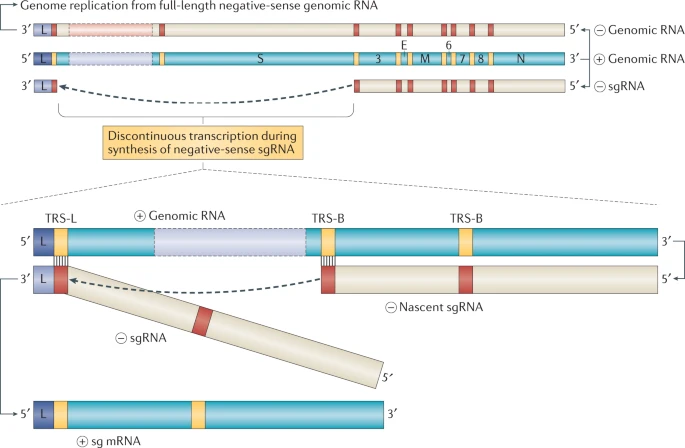
SARS-Cov-2 replication naturally use a complex mechanism of discontinuous transcription that involve a template switch between two different regions of the SARS-2 genome https://www.nature.com/articles/s41579-020-00468-6
This is not hard to imagine that errors in template switching would cause the replication complex to jump from SARS-2 to human mRNA, creating chimeric RNA sequences
The good thing is that hypothesis is very easy to test: I predict that all chimeric sequences will originate from exons (or other transcribed regions), and that they will be very few (or none) chimeric reads from introns or intragenic sequences
If these chimeric reads originated from integrated SARS-2 sequences, then you would expect them to originate mostly from intra- or inter-genic regions, and very rarely from exons. The opposite being true if these reads comes from template switching errors
The fact that the authors didn't mention and test this possibility tells me that they understand very little about SARS-2 biology, and even worse, that they didn't even try
(And this is assuming that these reads are not mere sequencing artifacts, which they probably are on second thoughts)

 Read on Twitter
Read on Twitter