This is the Final nail in the coffin of the current qPCR testing strategy.
We have to get smarter about what we are targeting and stop harassing asymptomatic people.
Now, let’s imagine if these genome integration events can seasonally express.
Permanent PCR Purgatory https://twitter.com/biorxiv_genomic/status/1338235108936052738
We have to get smarter about what we are targeting and stop harassing asymptomatic people.
Now, let’s imagine if these genome integration events can seasonally express.
Permanent PCR Purgatory https://twitter.com/biorxiv_genomic/status/1338235108936052738
Looks like the higher the copy number Subgenomic RNA, the higher frequency of human genomic integration.
Testing with the N gene is a disaster.
Testing with anything that subgenomic is a disaster.
Testing with the N gene is a disaster.
Testing with anything that subgenomic is a disaster.
This is nuts!
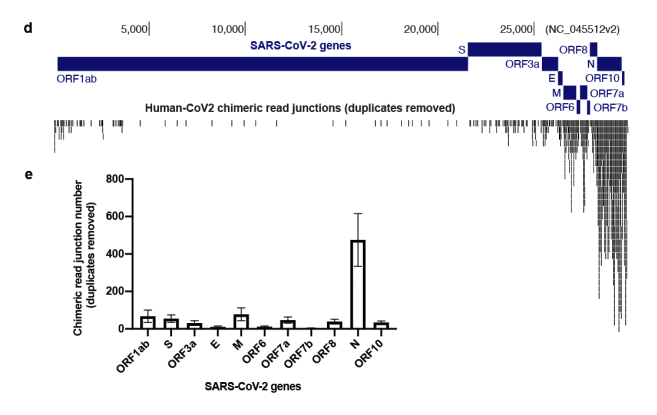
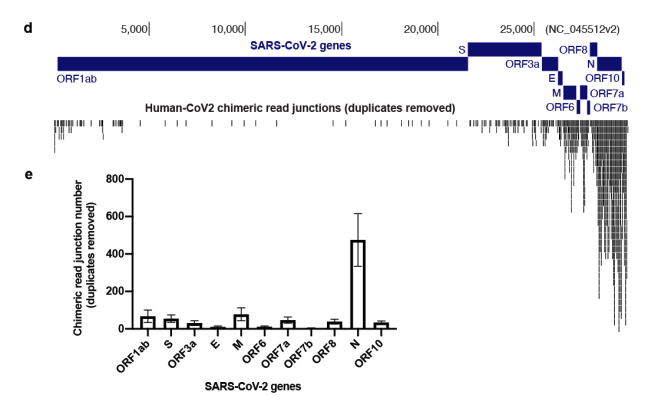
The integration is most common for the N gene.
That’s the most unfortunate news as most qPCR kits target the N gene.
They demonstrate the integration events can be expressed.
Stress can induce LINE-1 expression.
@PatrickSSte
The integration is most common for the N gene.
That’s the most unfortunate news as most qPCR kits target the N gene.
They demonstrate the integration events can be expressed.
Stress can induce LINE-1 expression.
@PatrickSSte
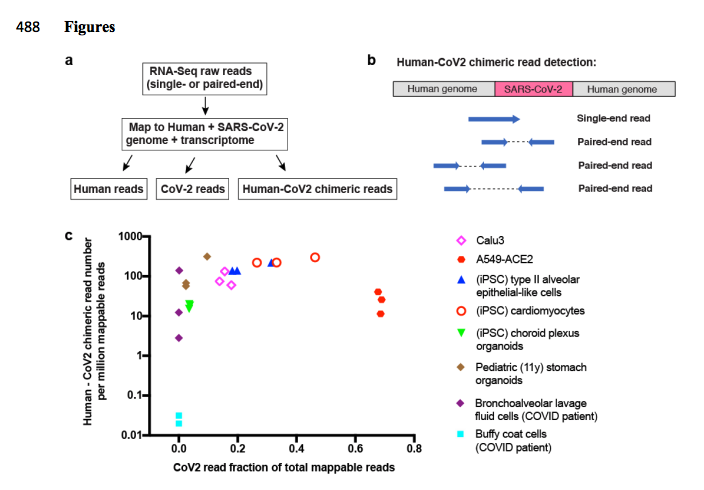
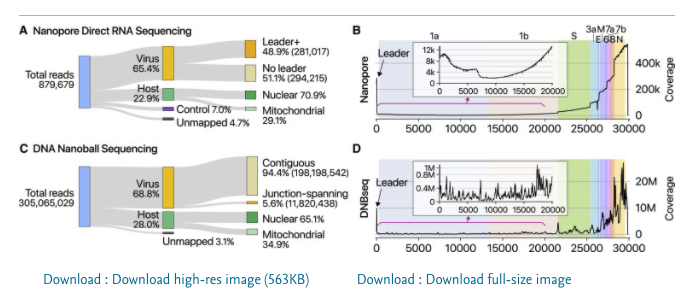
On the left is a reads/million chart across various cells lines and samples 100- 1000 reads/million. qPCR will pick this up as a 1:1000- 1:10,000 dilution of viral background. 9 Cq shift. Would explain the long clearance time on qPCR as your epithelial cells need to turn over.
It does it most with subgenomic RNAs and the most expressed one is the N gene. Also happens to be the most popular target for qPCR testing.
We really need ORF1a amplicon data to compare this to.
We really need ORF1a amplicon data to compare this to.
It's not peer reviewed and people are going to want to investigate these reads as at these reads/millions Illumina cluster errors can make chimeric reads. The authors do comb over public SRA data and confirm the observations but the Seq data from the Preprint isn't live yet.
qPCR positivity in stool is the longest but very few groups get the stool samples to plate. I would have though those cells turn over fastest so this needs more digging.
Permanent PCR Purgatory is hyperbole
Prolonged PCR Purgatory is fair description of the problem.
Prolonged PCR Purgatory is fair description of the problem.
One of the push backs on this will be the chimeric read rate in Illumina sequencers. This is real but I just dont see how that would match the biology of the subgenomicRNAs.
They see the most integrations events with the most highly expressed subgenomic RNA. Kim et al
They see the most integrations events with the most highly expressed subgenomic RNA. Kim et al
Compare that to their map of chimeric junctions seen in the inverted SARs-CoV-2 genome histogram (top). Random chimeric reads combined with the LINE-1 data and induction data. If I had to guess without the seqdata.. I bet its real.
In order for you to fully clear qPCR testing you need to wait for every cell it has infected/integrated with to die and turnover. Epithelial cells do this faster than most.
English translation: We are not targeting active replicating virus. We are targeting it's Ghost.
English translation: We are not targeting active replicating virus. We are targeting it's Ghost.
How long that Ghost haunts you is seen in many papers @carlheneghan has reviewed. If it infects any stem cells, their will be progeny which may take longer to clear. https://www.medrxiv.org/content/10.1101/2020.08.04.20167932v4
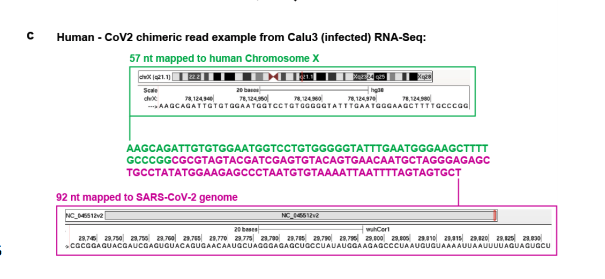
N example of a chimeric read.
What constitutes perfect evidence for this? The good news is everyone will start paying attention to the impact of subgenomicRNA on false qPCR signals. N amplicons over-count non-infectious viral RNA whether they live in DMVs or chromosomes
What constitutes perfect evidence for this? The good news is everyone will start paying attention to the impact of subgenomicRNA on false qPCR signals. N amplicons over-count non-infectious viral RNA whether they live in DMVs or chromosomes

 Read on Twitter
Read on Twitter