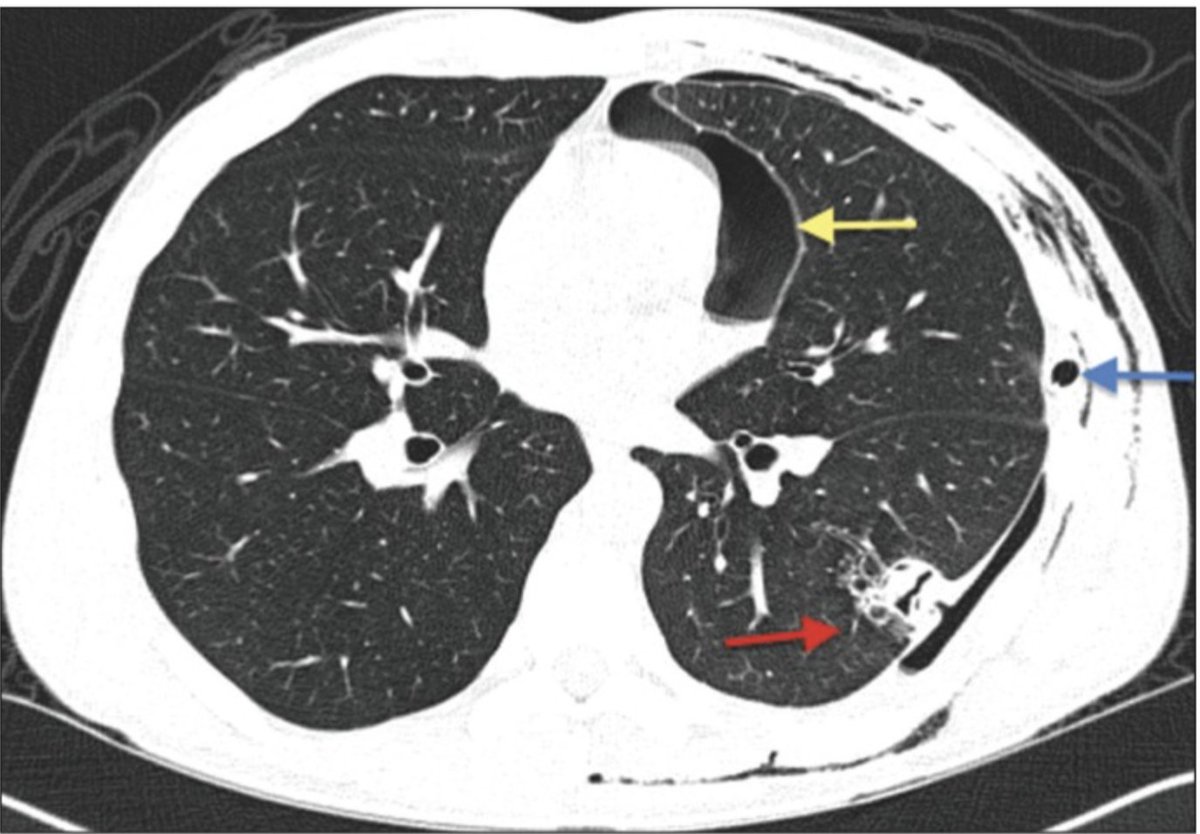
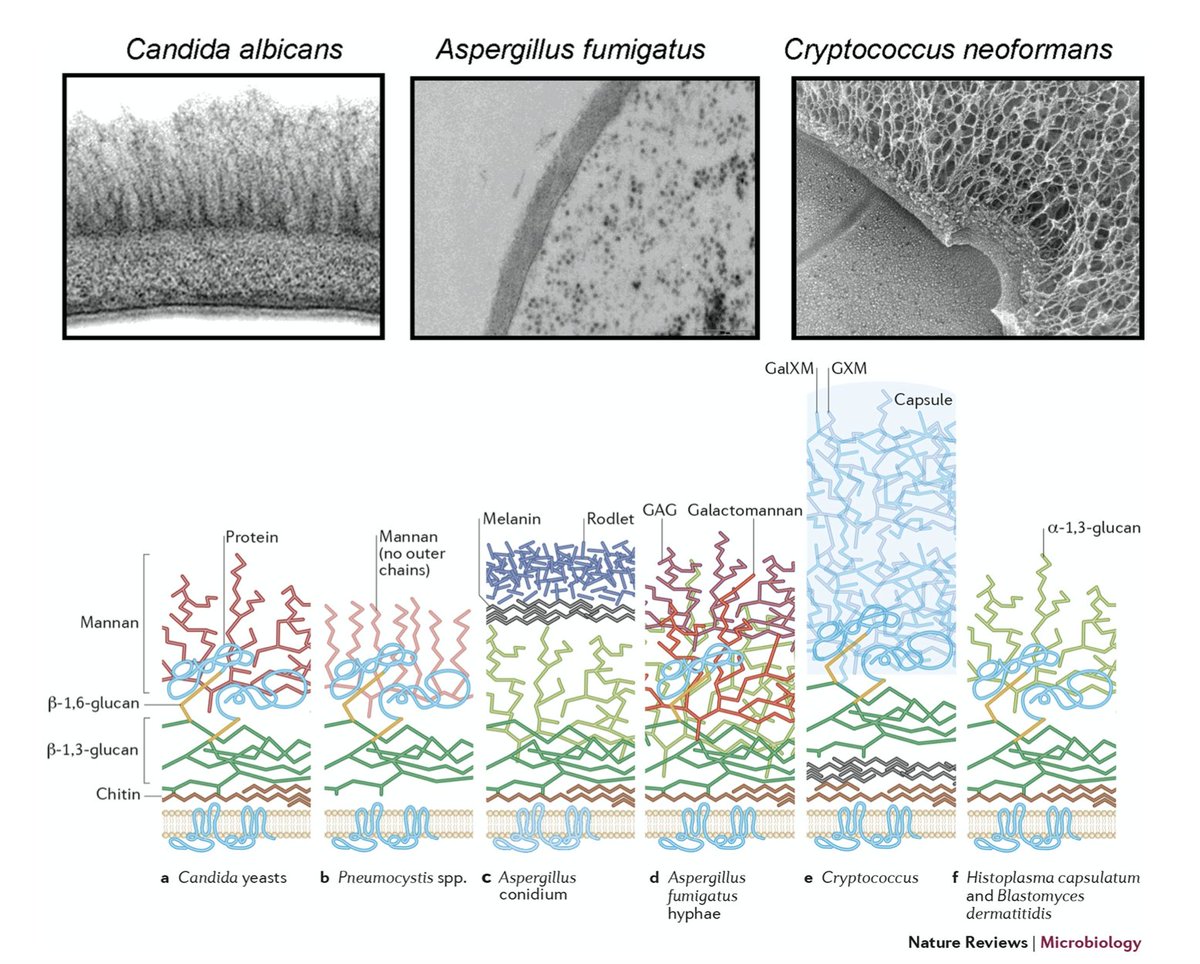
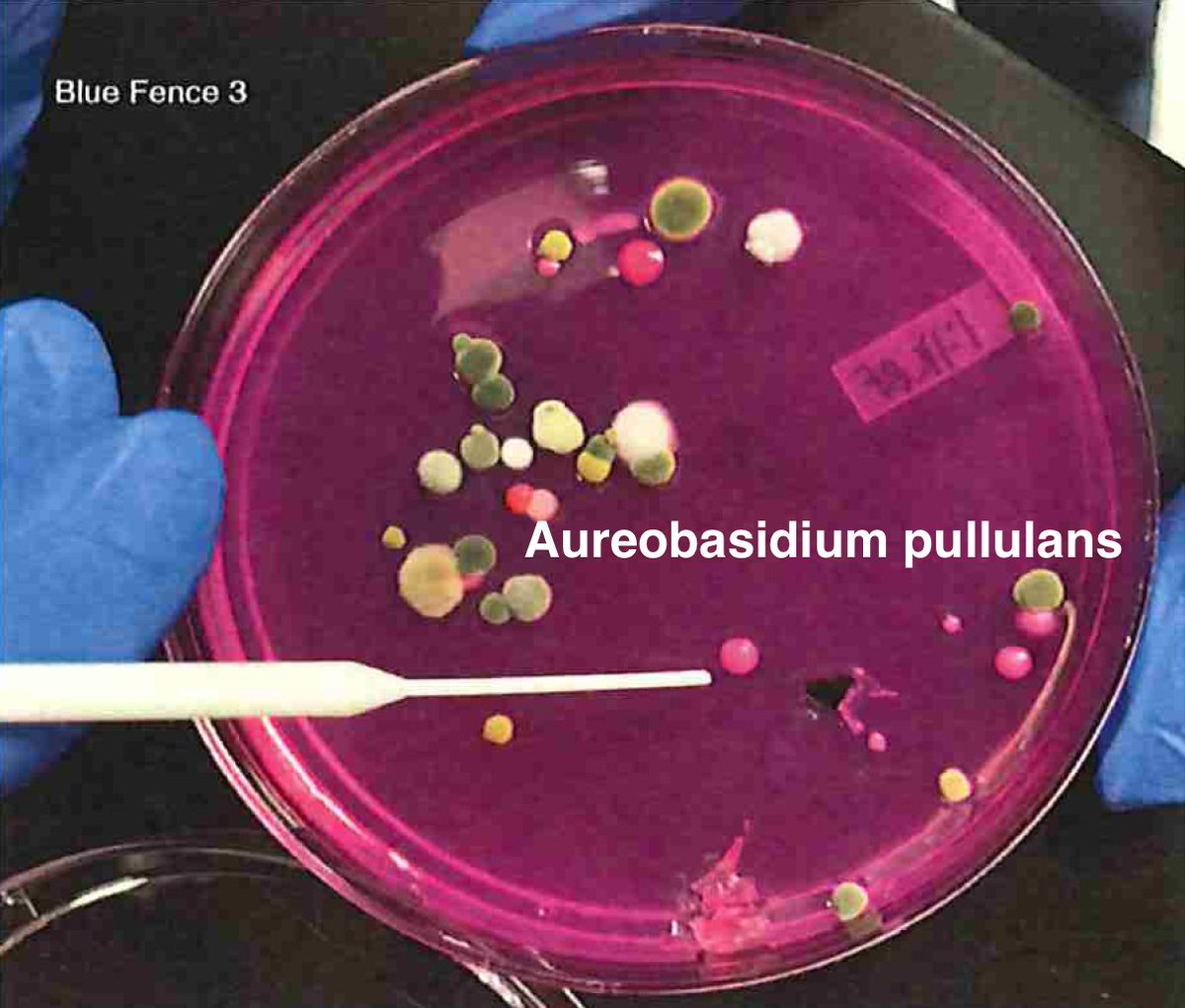
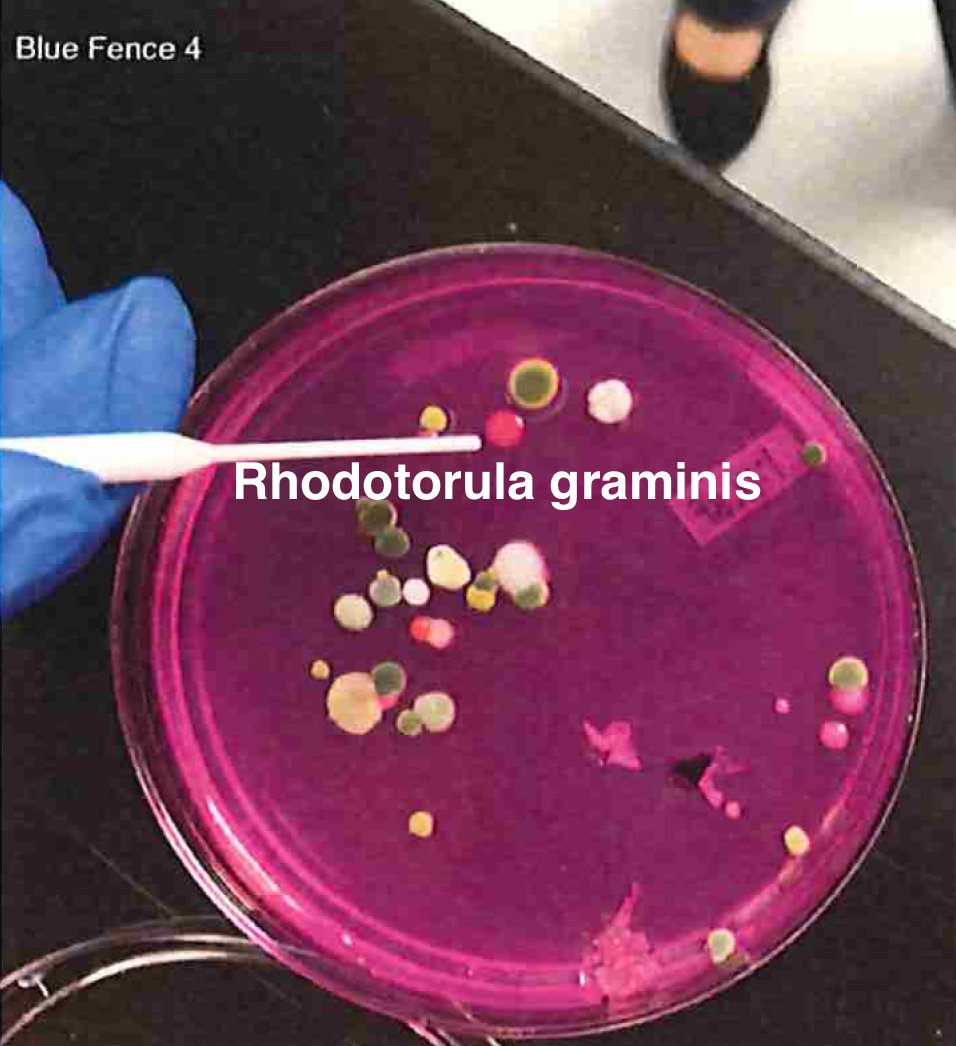
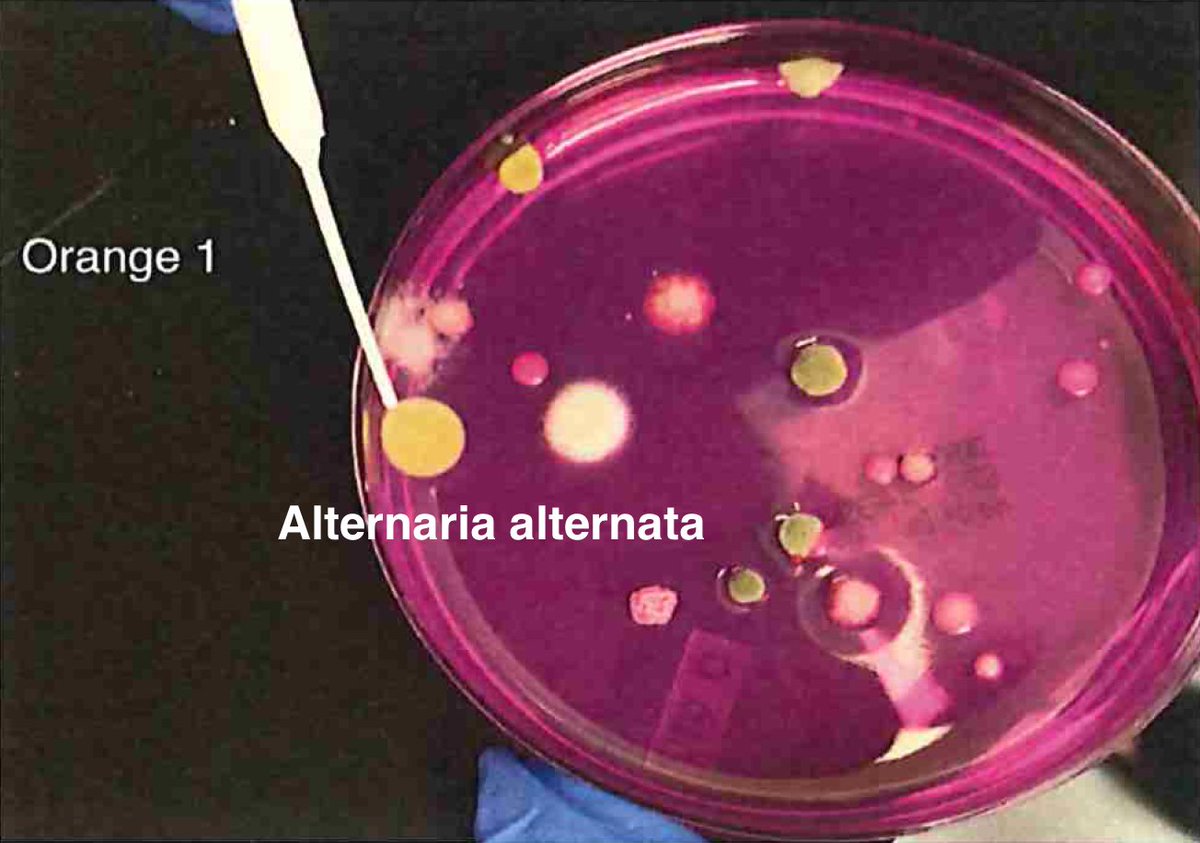
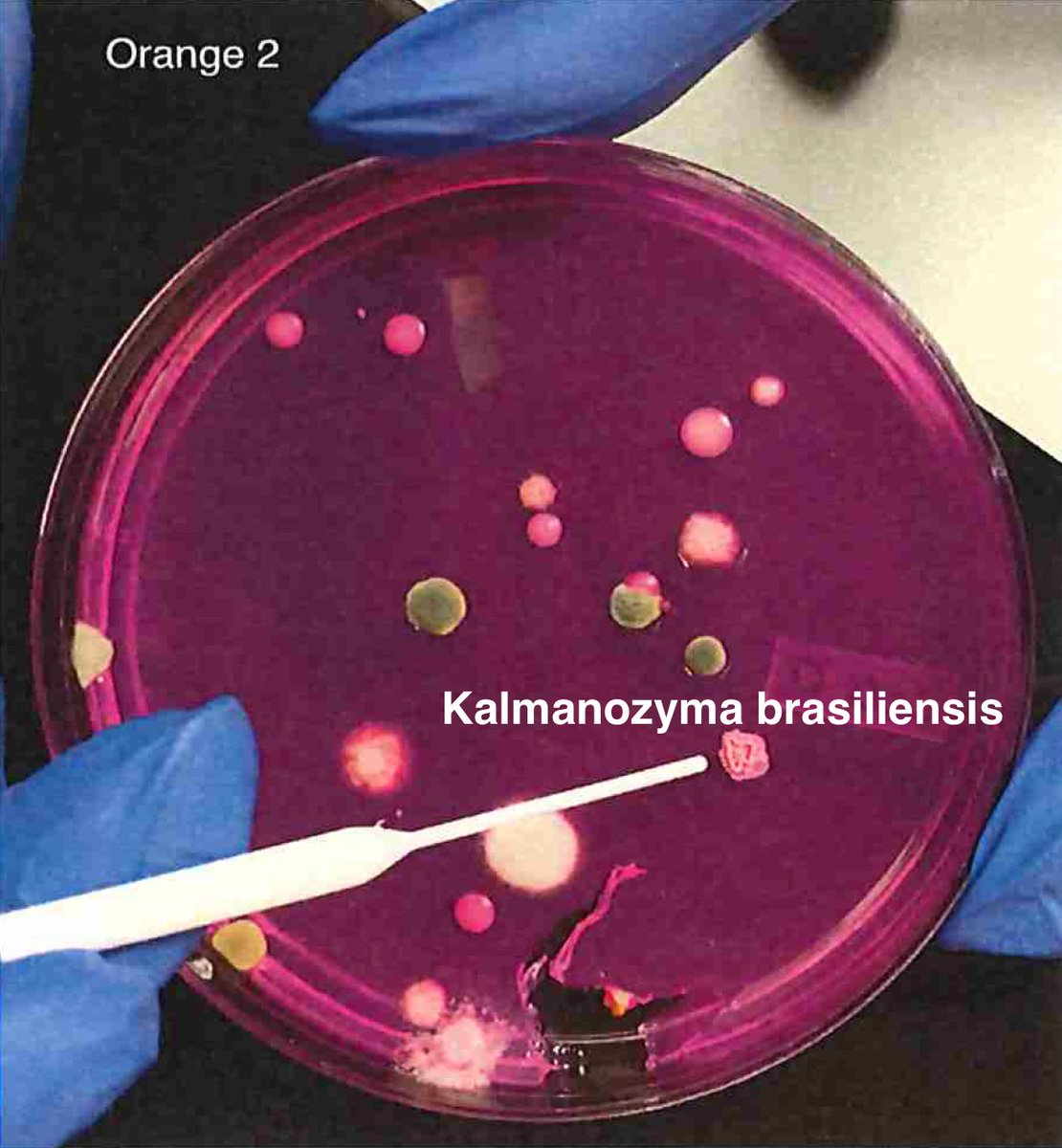
Let's walk through the much more complicated problem of identifying pathogens on Cannabis flower. These are inhaled products in an industry that just experienced EVALI.
This will provide a few examples of which steps in the process are prone to producing illusions with qPCR.
This will provide a few examples of which steps in the process are prone to producing illusions with qPCR.
The first problem is that Cannabis microbes can be both Endophytes and Epiphytes. These live inside the plant or on the surface of the plant. This changes how you survey them. If you don't lyse open plant cell walls, you cant see the Endophytes. You need proof of lysis.
We accomplish this by targeting a conserved cannabis sequence in qPCR. Every PCR assay must have a qPCR target that proves you lysed open the plant cells.
Many SARs-CoV-2 assays accomplish this by targeting a human gene known as RNaseP.
Many SARs-CoV-2 assays accomplish this by targeting a human gene known as RNaseP.
If you don't have this, you have no proof of lysis and can only measure the Epiphytes.
However, it's the Endophytes like Aspergillus that are responsible for the only clinical fatalities in the cannabis space. People don't overdose on THC. But Aspergillosis is serious.
However, it's the Endophytes like Aspergillus that are responsible for the only clinical fatalities in the cannabis space. People don't overdose on THC. But Aspergillosis is serious.
Once you know you can lyse open cannabis, you need to know if you can lyse open all Yeast and Molds.
This is not so trivial as fungi have cell membranes coated in cell walls with thick Chitin and Glucan coats.
This is not so trivial as fungi have cell membranes coated in cell walls with thick Chitin and Glucan coats.
Adding insult to injury, Cannabis flowers are dried in a curing process which helps to desiccate the bacteria but stimulates many yeast and molds to thicken their cell walls making them harder to lyse. To get qPCR to correlate with viable yeast and molds, we need Live-Dead tricks
A good review on this problem is published here.
https://pdfs.semanticscholar.org/c523/06946e661bc54af90cf4d58b96766f9d6fa9.pdf
https://pdfs.semanticscholar.org/c523/06946e661bc54af90cf4d58b96766f9d6fa9.pdf
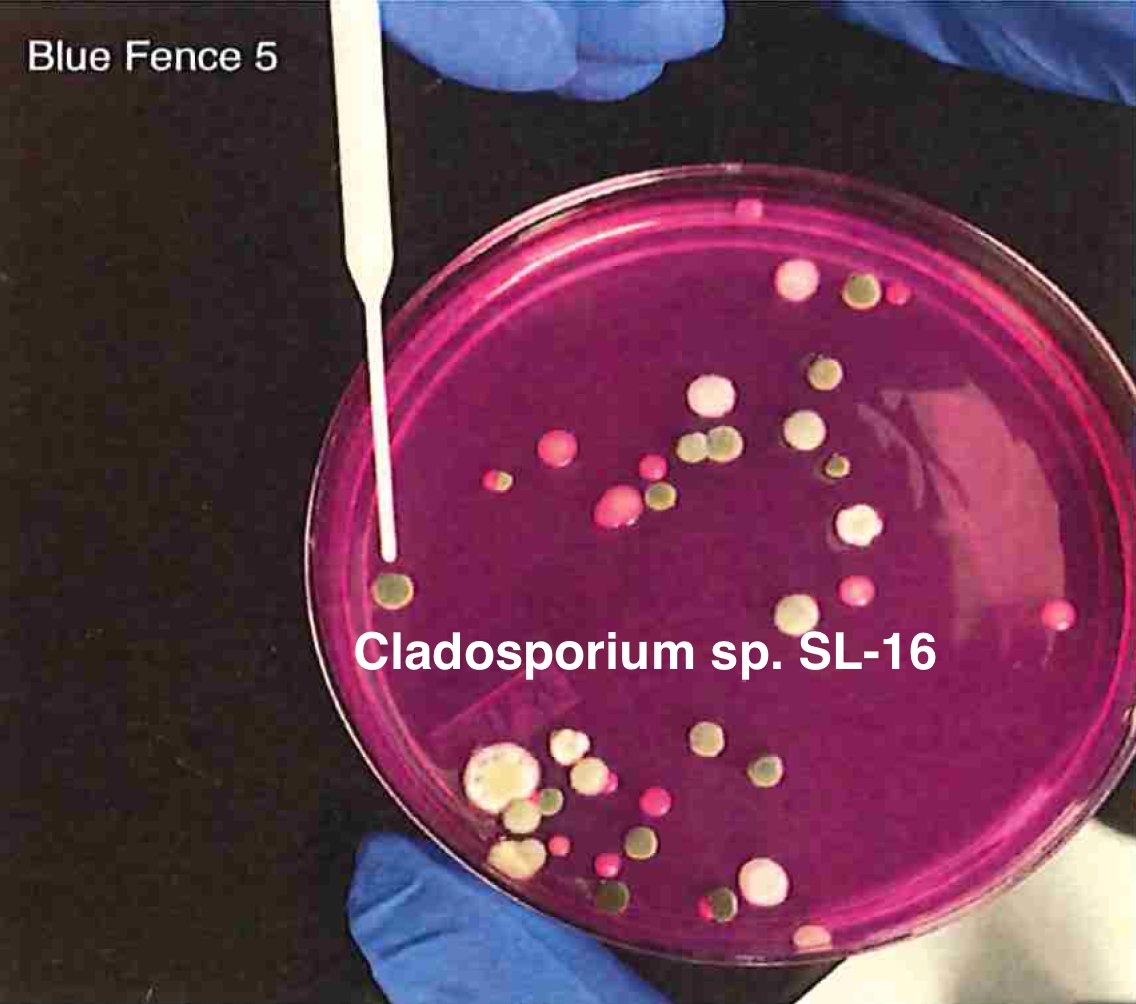
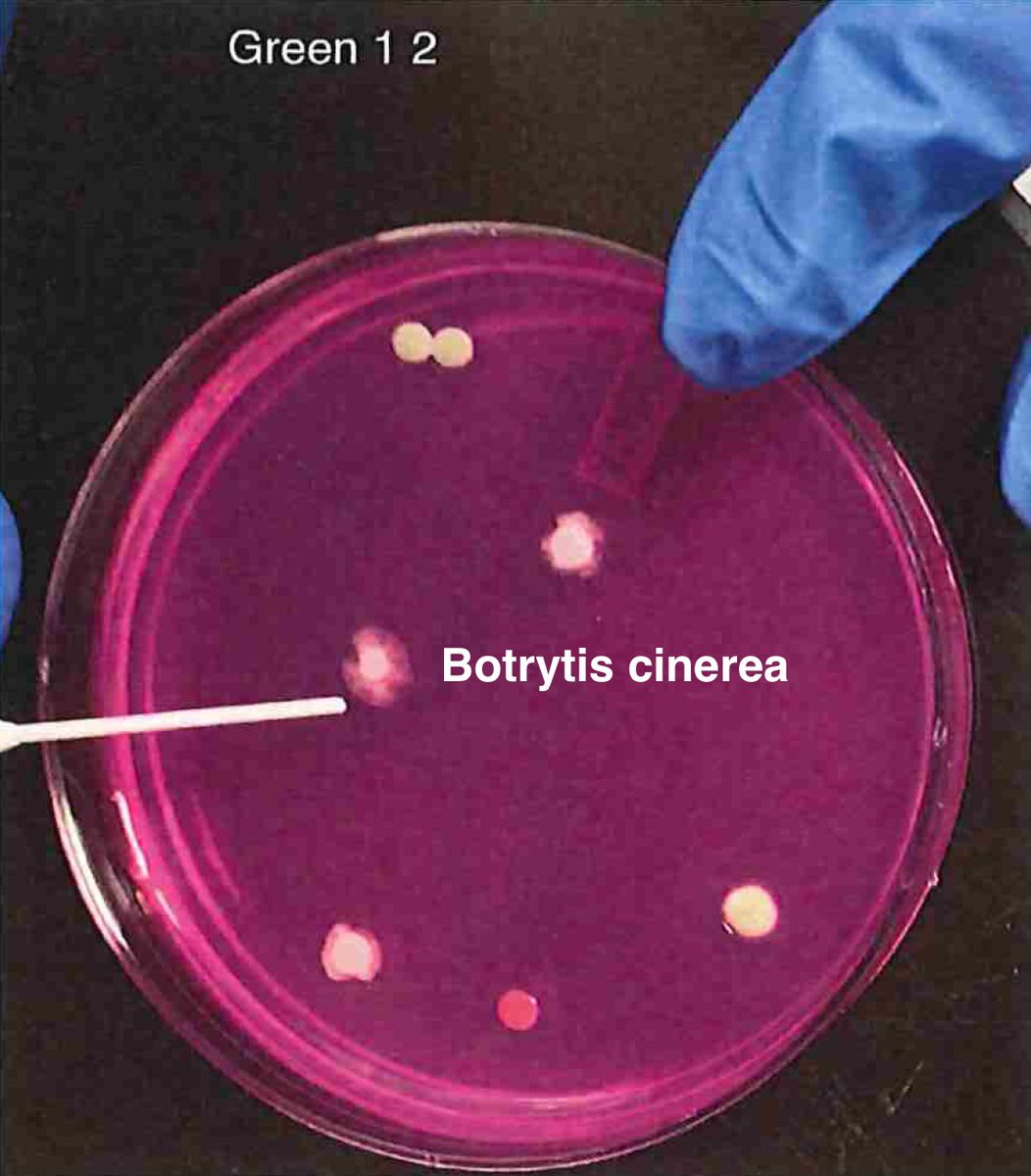
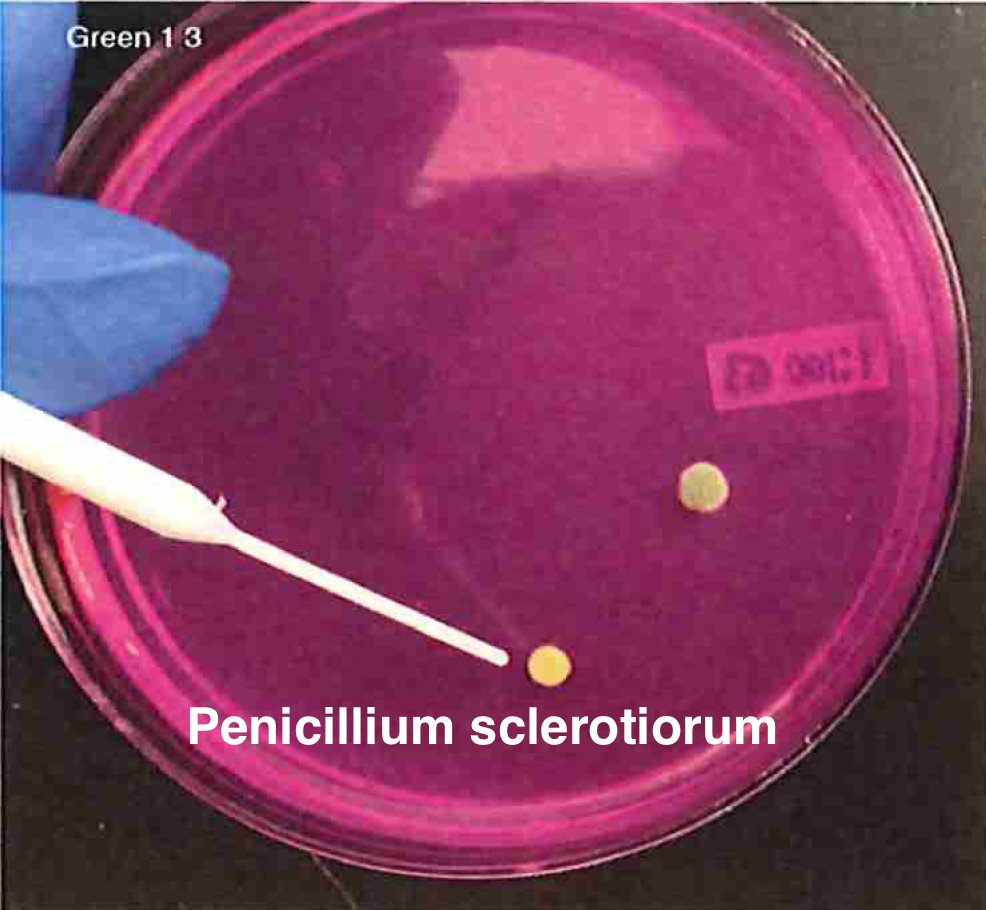
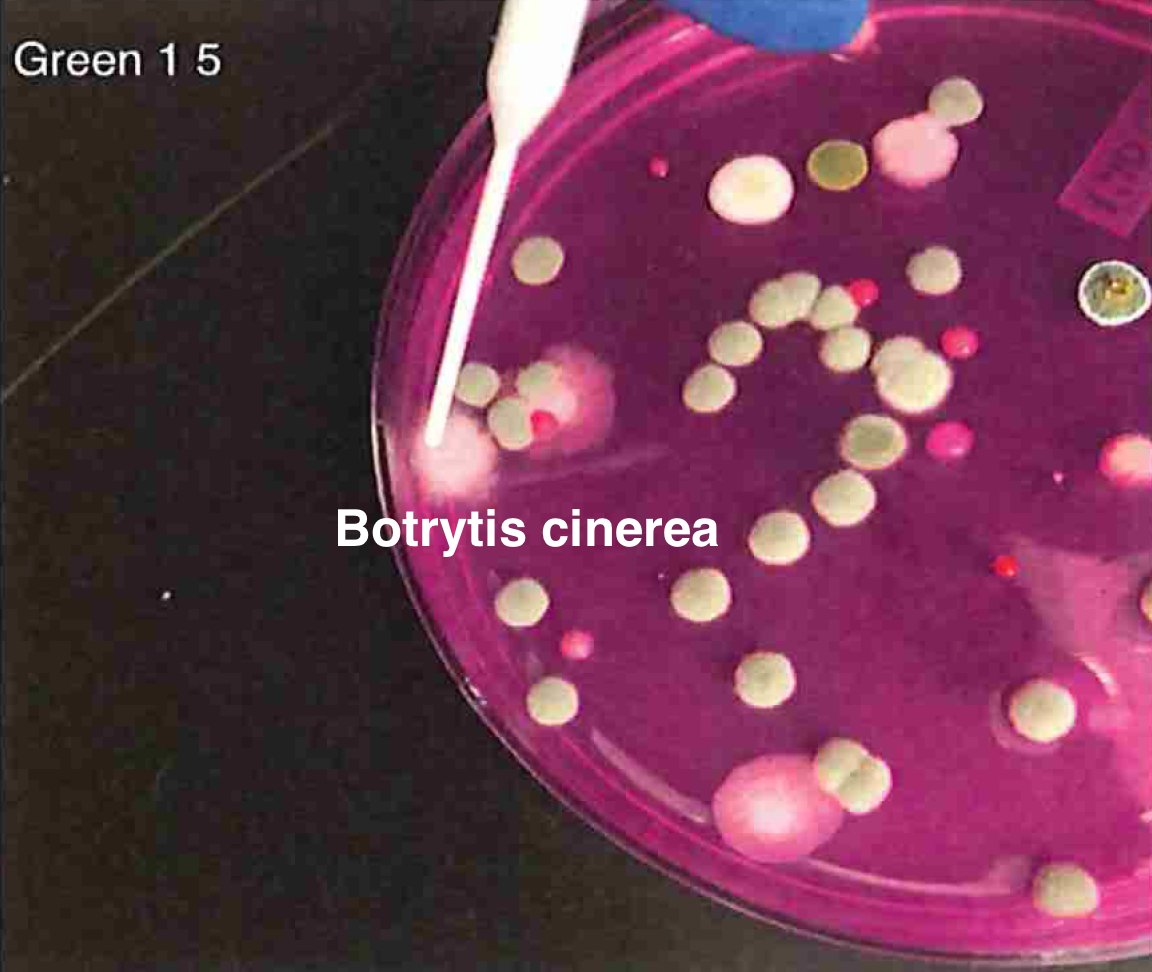
So unlike some people in the rush to make millions in qPCR testing off of C19, we first plate the microbes to understand the diversity of microbes we are dealing with and how many are viable. This is needed as the curing process kills some bacteria but not all the fungi.
We then measure the CFU's from each type of microbe and how that correlates with ITS qPCR Signals. This is analogous to Jaafar et al. work on C19 on Vero cells.
Note, this was NOT done with the C19 protocol at the WHO.
These orgs were acquired from reputable biobanks -ATCC
Note, this was NOT done with the C19 protocol at the WHO.
These orgs were acquired from reputable biobanks -ATCC
Once we have an understanding of how ideal controls perform regarding viability in culture, we turn to real biological samples where the microbiome is unknown.
We are going to need whole genome sequencing to sort this out. Not PCR as we cant afford the primer bias.
We are going to need whole genome sequencing to sort this out. Not PCR as we cant afford the primer bias.
What do I mean by primer bias?
Your primers can only amplify organisms that match your primers and its possible any given selection of ITS primers will miss certain microbes with mutations under the primers.
Whole Genome Sequencing is unbiased in this regard.
Your primers can only amplify organisms that match your primers and its possible any given selection of ITS primers will miss certain microbes with mutations under the primers.
Whole Genome Sequencing is unbiased in this regard.
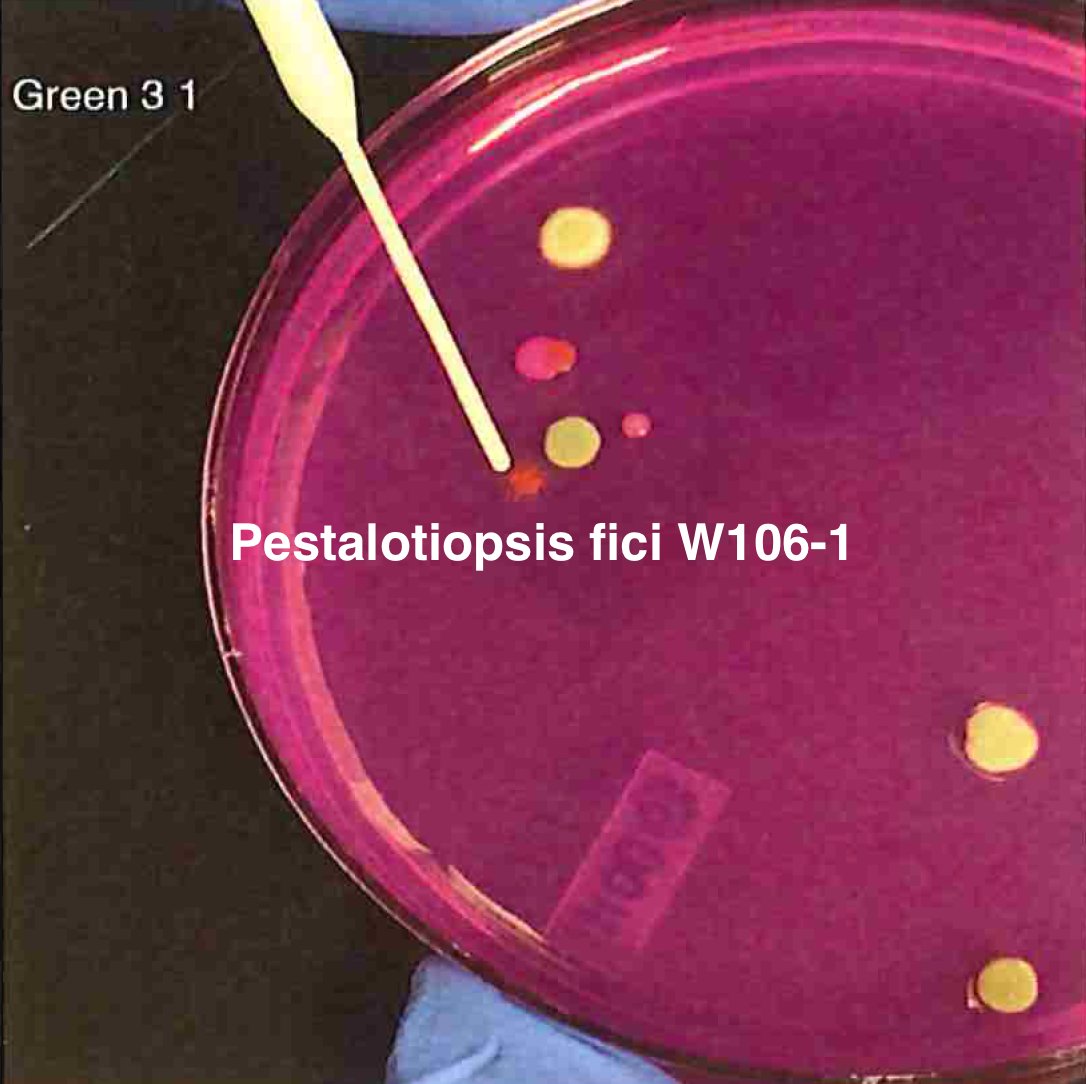
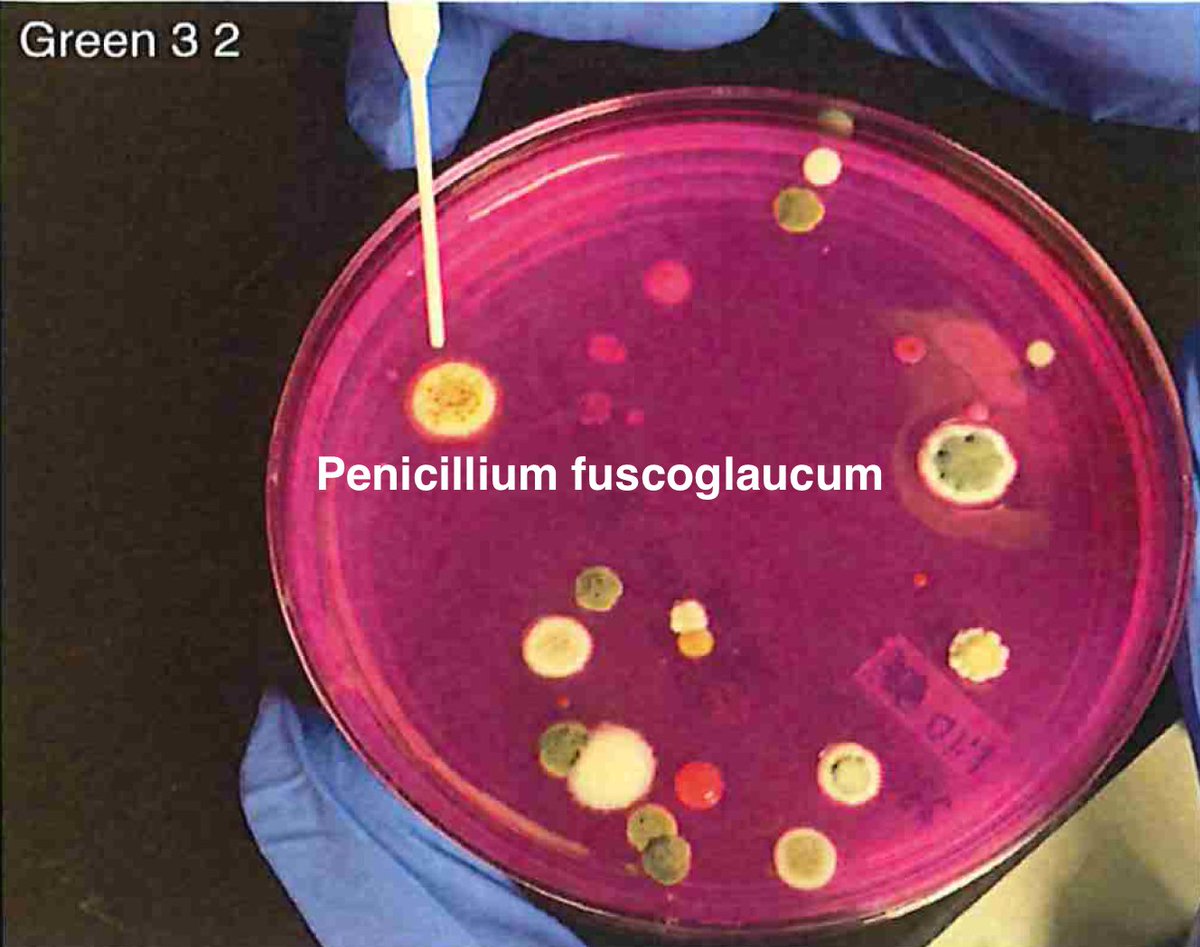
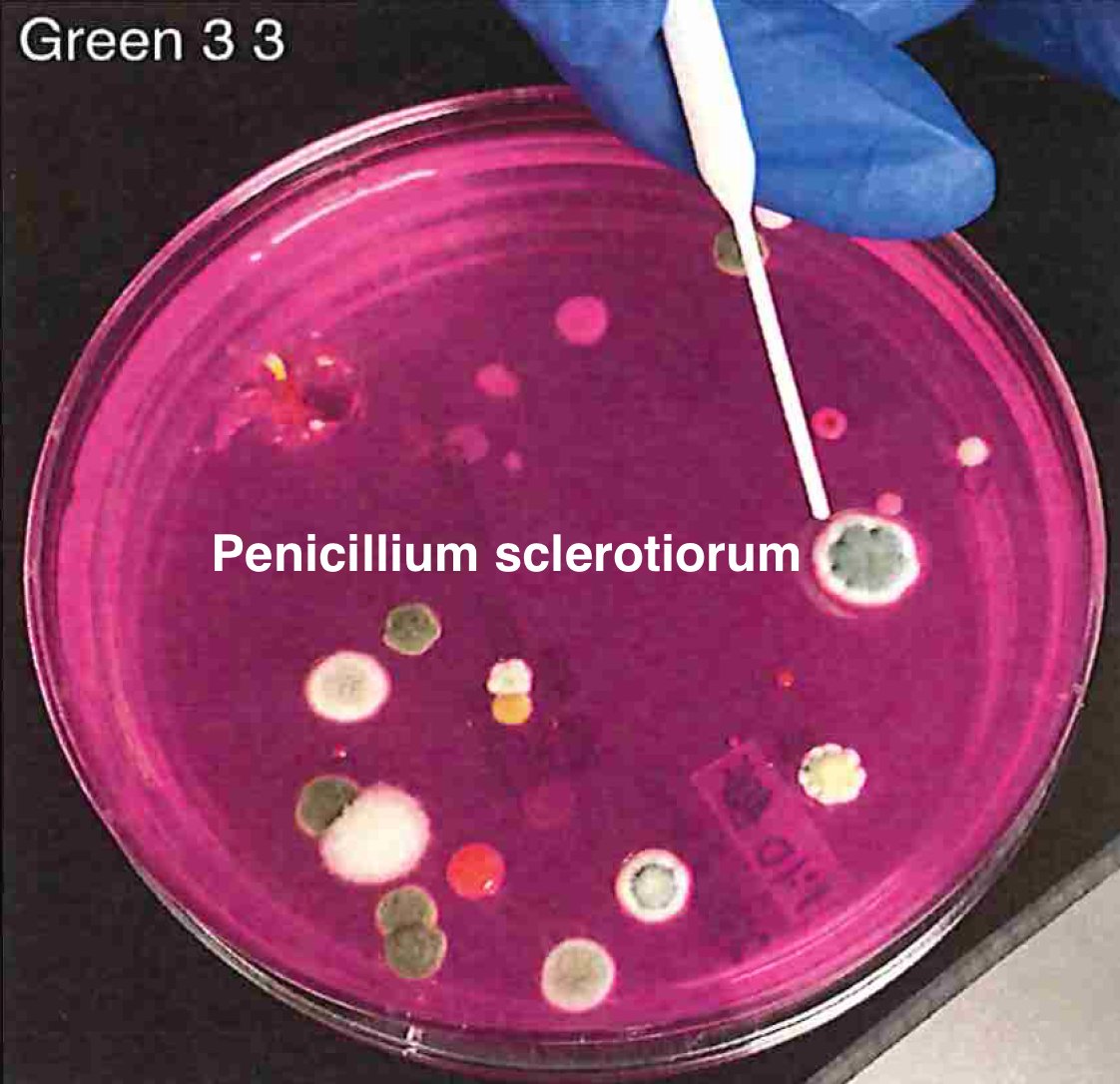
So plate microbiomes from 14 different outdoor cannabis plants and begin to isolate each pathogen and sequence its genome. Part of this isolation process is ensuring our Lysis conditions liberate DNA. DNA coming from culture only represents the DNA that is in viable organisms.
In practice, our PCR assay will face both live and dead DNA and the culture methods will only count live DNA. We have a solution for this which we will touch on later.
So we isolate colonies, understand their plating curves and how they correlate with qPCR and how well they lyse
So we isolate colonies, understand their plating curves and how they correlate with qPCR and how well they lyse
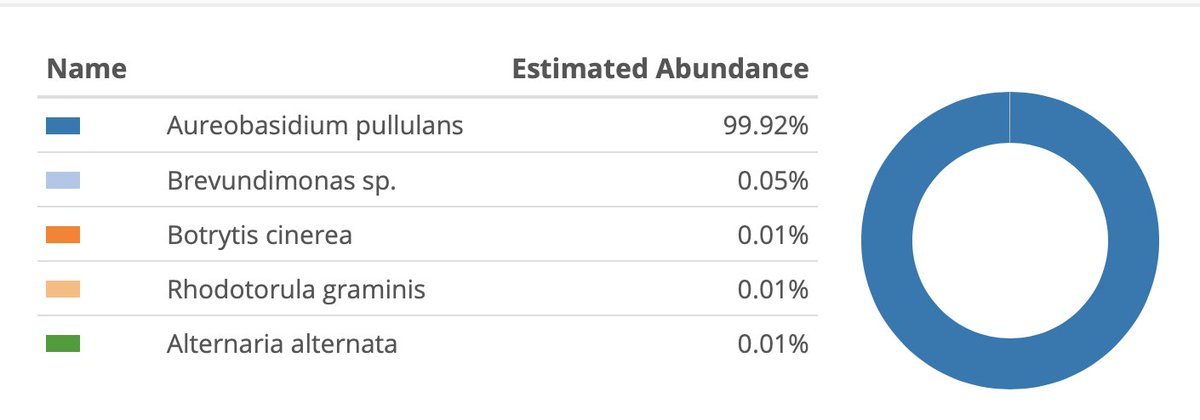
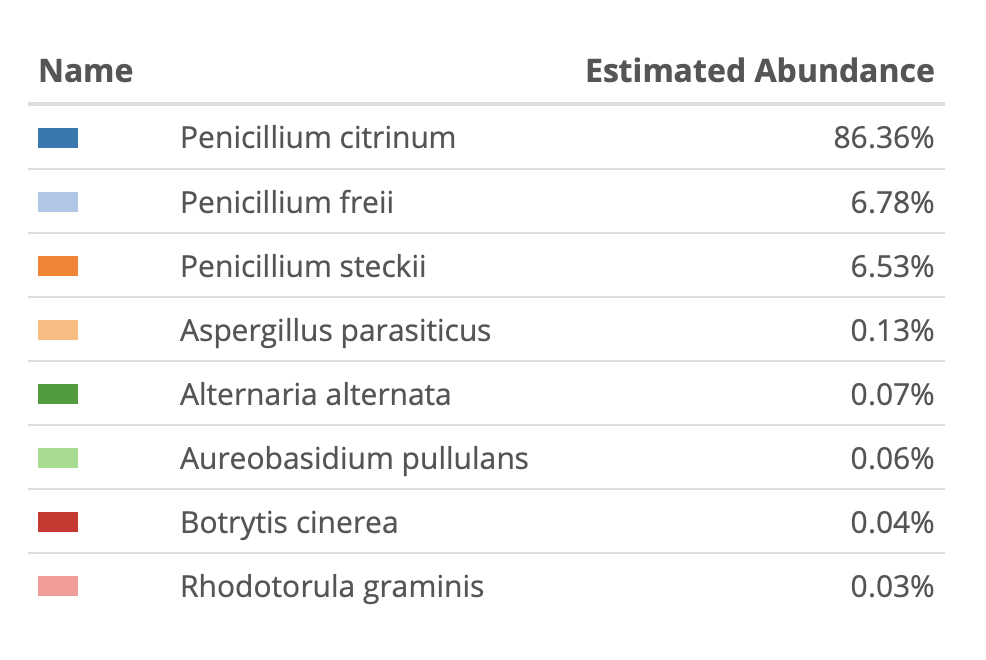
Then we move to Whole Genome Sequencing. We subsample 1 Million Illumina reads and as expected most genomes come back pure.
BF3= https://app.onecodex.com/analysis/public/a6e9f1f5a33f4edd
BF4 = https://app.onecodex.com/analysis/public/6dda9e399ed14216
BF5 = https://app.onecodex.com/analysis/public/476495de3fc04565
G1-2 = https://app.onecodex.com/analysis/public/7bcdd96875a44496
G1-3 = https://app.onecodex.com/analysis/public/94b4d67e02f647df
BF3= https://app.onecodex.com/analysis/public/a6e9f1f5a33f4edd
BF4 = https://app.onecodex.com/analysis/public/6dda9e399ed14216
BF5 = https://app.onecodex.com/analysis/public/476495de3fc04565
G1-2 = https://app.onecodex.com/analysis/public/7bcdd96875a44496
G1-3 = https://app.onecodex.com/analysis/public/94b4d67e02f647df
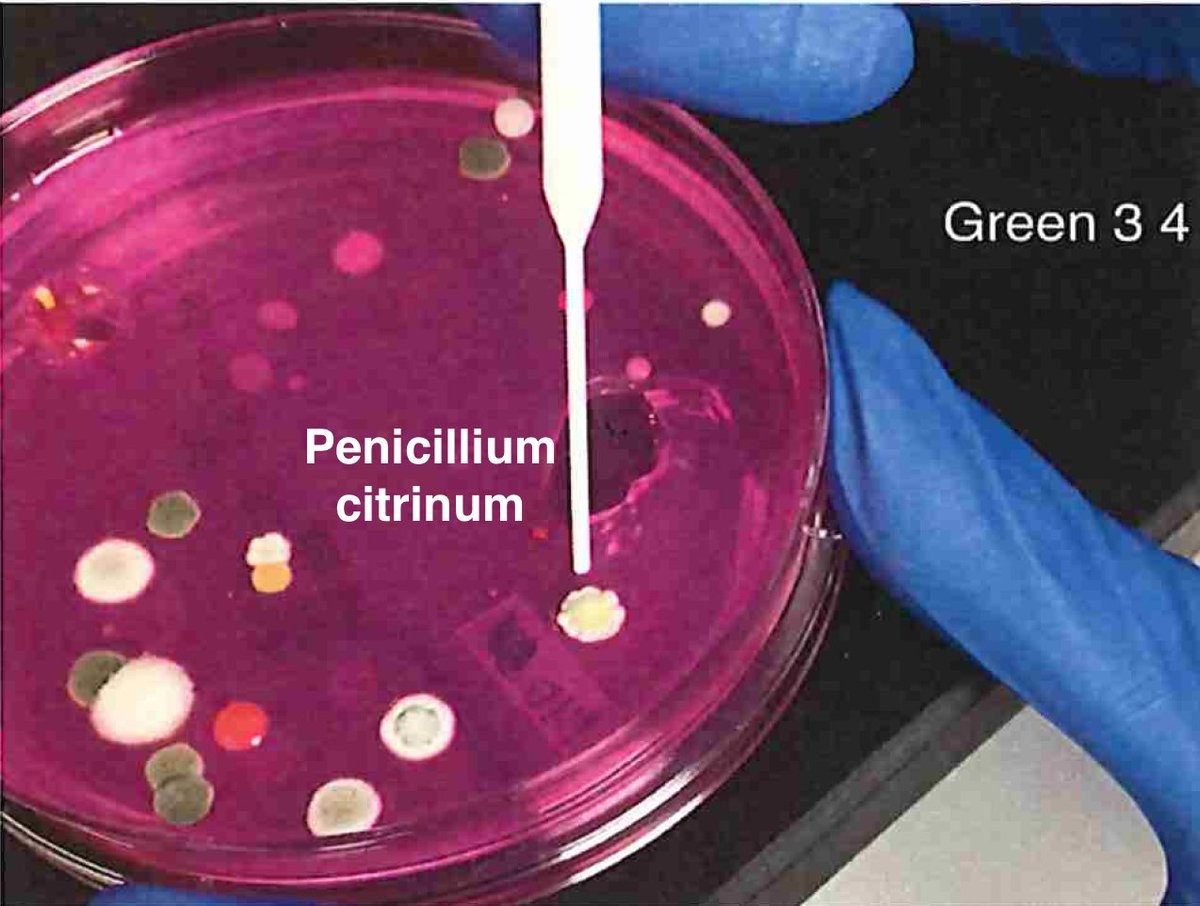
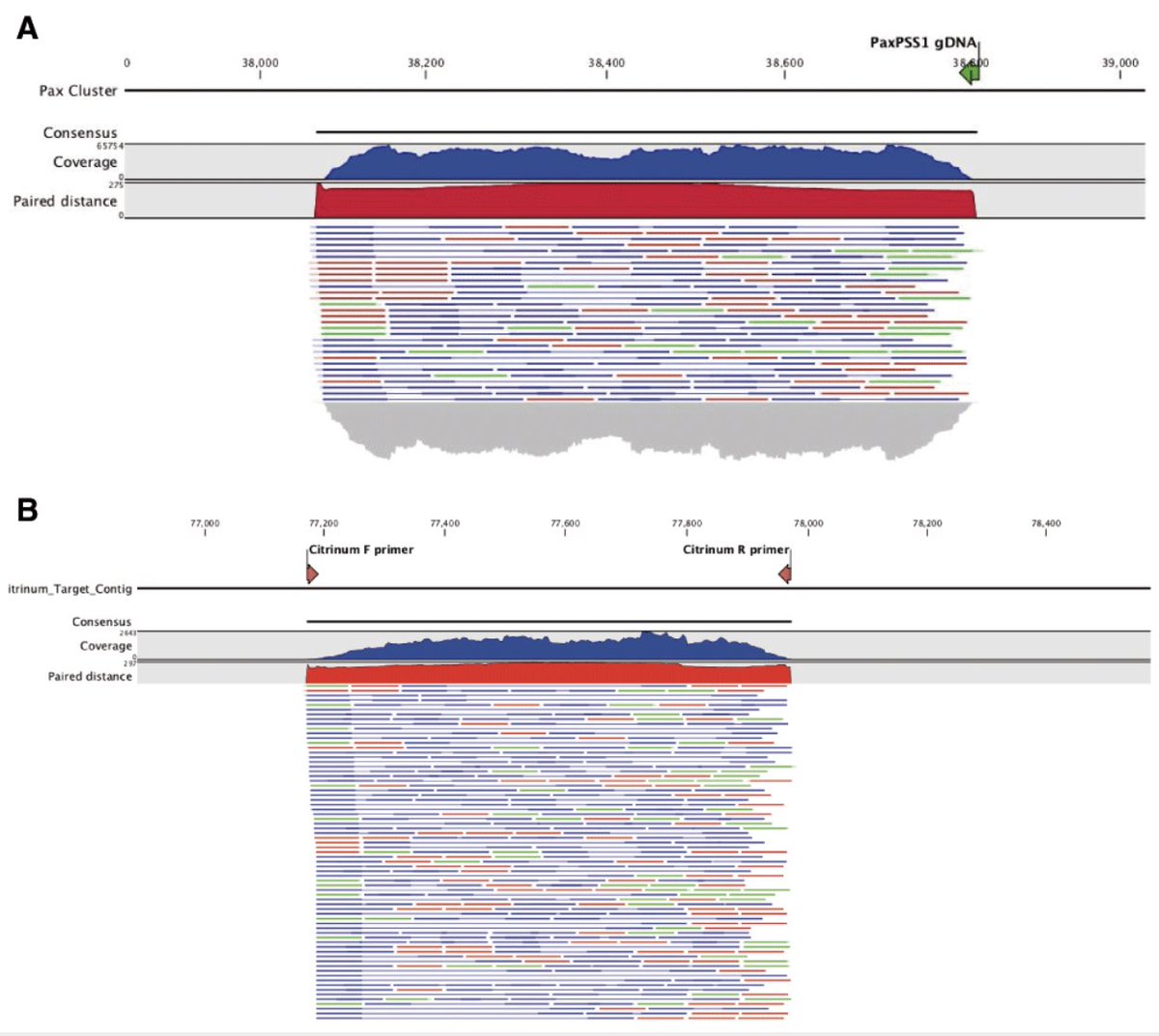
Now we can confirm our primer sites exist in the biological targets. P.citrinum we see often.
G1-4 = https://app.onecodex.com/analysis/public/987449aec4c34abc
G1-5 = https://app.onecodex.com/analysis/public/3c69faf6f6084010
G3-1= https://app.onecodex.com/analysis/public/ac1c01792d3e4088
G3-2 = https://app.onecodex.com/analysis/public/98bf391c12674297
G3-3 = https://app.onecodex.com/analysis/public/385b829b1ff641d3
G3-4 = https://app.onecodex.com/analysis/public/a7c93912ccb2488c
G1-4 = https://app.onecodex.com/analysis/public/987449aec4c34abc
G1-5 = https://app.onecodex.com/analysis/public/3c69faf6f6084010
G3-1= https://app.onecodex.com/analysis/public/ac1c01792d3e4088
G3-2 = https://app.onecodex.com/analysis/public/98bf391c12674297
G3-3 = https://app.onecodex.com/analysis/public/385b829b1ff641d3
G3-4 = https://app.onecodex.com/analysis/public/a7c93912ccb2488c
We see different microbiome from varietals, B, G and O.
Penicillium citrinum we have published on this before...
In an open review journal. The Review is public.
O1 = https://app.onecodex.com/analysis/public/b765b329741f459f
O2 = https://app.onecodex.com/analysis/public/aaef7f5b18e24063
O3 = https://app.onecodex.com/analysis/public/e05f574ffd7e40b6
https://f1000research.com/articles/4-1422
Penicillium citrinum we have published on this before...
In an open review journal. The Review is public.
O1 = https://app.onecodex.com/analysis/public/b765b329741f459f
O2 = https://app.onecodex.com/analysis/public/aaef7f5b18e24063
O3 = https://app.onecodex.com/analysis/public/e05f574ffd7e40b6
https://f1000research.com/articles/4-1422
You'll notice that when we get a qPCR positive sample, we don't just assume it's on target.
We sequence the amplicons to be sure and we even design a second confirmatory amplicon that targets the toxin gene to prove its mycotoxin capable.
The WHO qPCR protocol skipped this step
We sequence the amplicons to be sure and we even design a second confirmatory amplicon that targets the toxin gene to prove its mycotoxin capable.
The WHO qPCR protocol skipped this step
One thing we learned in this process is that your lysis conditions can swing your Cq scores 5-8 Cqs! That an enormous variable that needs to be dealt with.
We learned a lesson from Powdery Mildew resistant cannabis to get this done. Some cannabis has figured out how to lyse mold
We learned a lesson from Powdery Mildew resistant cannabis to get this done. Some cannabis has figured out how to lyse mold
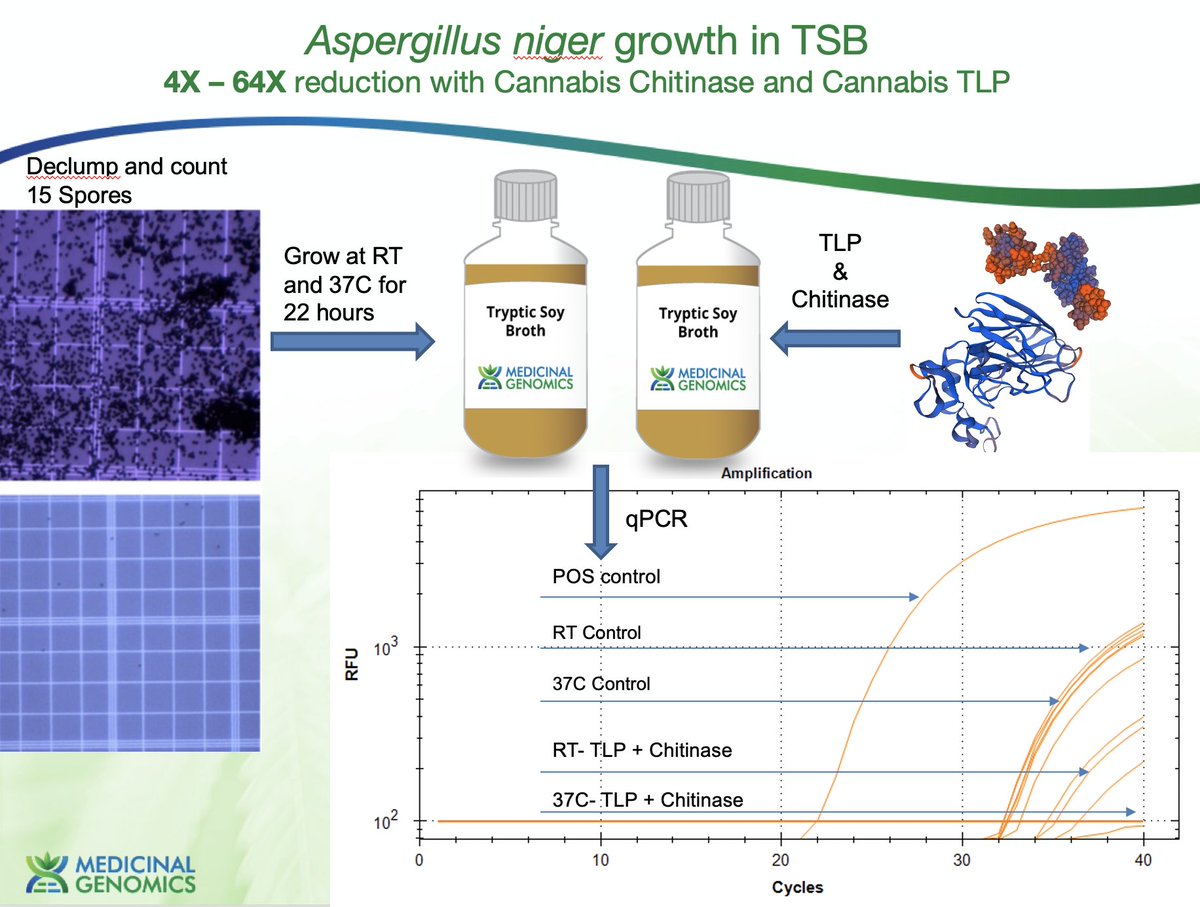
This required 4 Terabases of sequencing across 42 cannabis genomes to smoke out. It led us to clone a few Chitinases and Thaumatin-Like Proteins we found in PM resistant cannabis. These turned out to be excellent Fungal lysis enzymes.
https://www.biorxiv.org/content/10.1101/2020.01.03.894428v1
https://www.biorxiv.org/content/10.1101/2020.01.03.894428v1
So we homogenize the flower in TSB with these enzymes and we see 4-64X more DNA from the yeast and molds and get far better concordance with plating and qPCR.
All Seq data public.
No guessing what Cq scores mean on assays rushed to the WHO with questionable peer review.
All Seq data public.
No guessing what Cq scores mean on assays rushed to the WHO with questionable peer review.
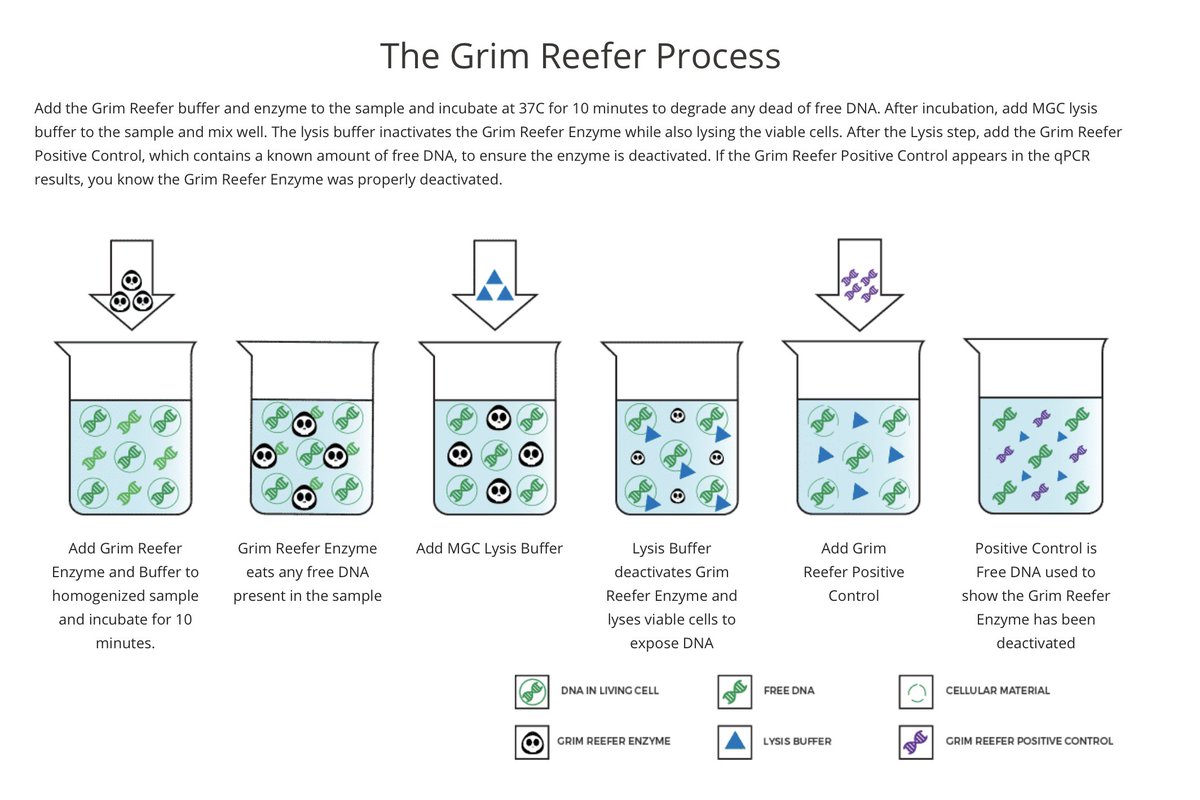
So how do we deal with Live-Dead. Surely there must be dead fungal DNA that makes qPCR over count microbes just like C19?
Well, we can play some tricks in the DNA world a little easier. Viable cells can be spun down and dead ones can be erased with nucleases.
Well, we can play some tricks in the DNA world a little easier. Viable cells can be spun down and dead ones can be erased with nucleases.
The process for doing this on fungi is a bit different than what we do for bacteria but the principle is the same. You need to separate live organisms from dead ones without waiting for them to grow for 5 days.
These microbes are homogenized in salty TSB.
These microbes are homogenized in salty TSB.
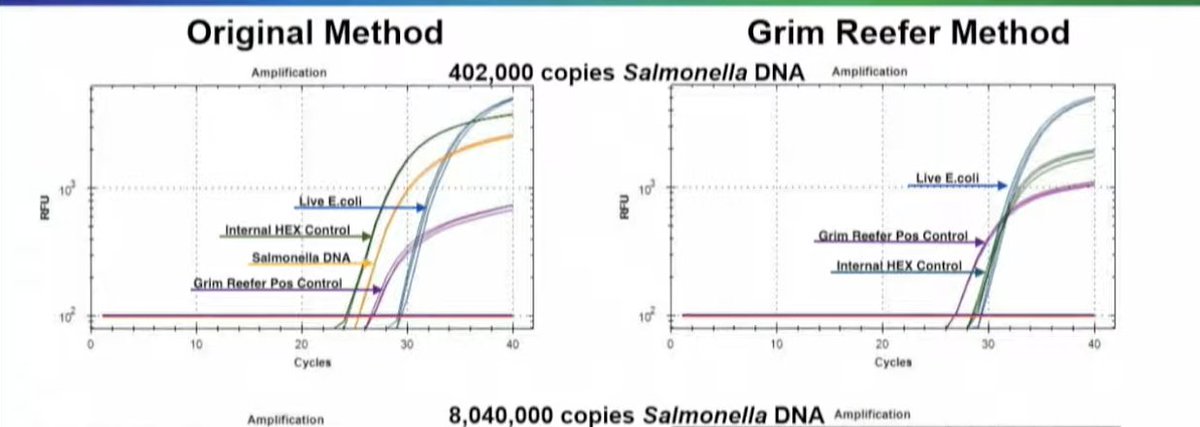
You look for nucleases that are salt tolerant and design another DNA control that you spike in after you kill these enzymes. This added control is in place to ensure your nucleases are infact dead before lysing open your cells.
We call this Grim Reefer- it brings out the dead.
We call this Grim Reefer- it brings out the dead.
Hopefully this gives you a sense of what's required to really know if PCR is measuring something biologically meaningful.
It's a very powerful tool that right now is pissing in the pool with sloppy C19 testing.
PCR shouldn't be a dirty word. It's miraculous.
It's a very powerful tool that right now is pissing in the pool with sloppy C19 testing.
PCR shouldn't be a dirty word. It's miraculous.
@threadreaderapp unroll

 Read on Twitter
Read on Twitter