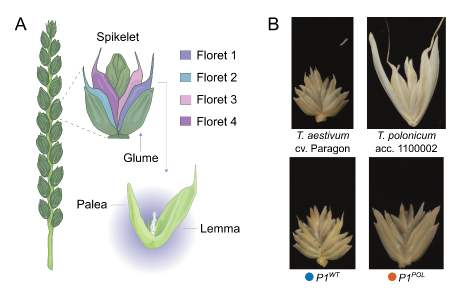
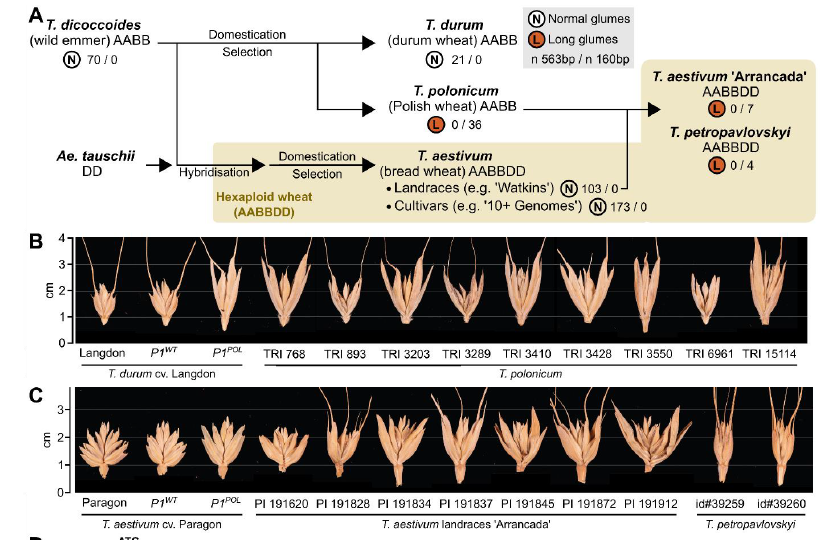
This is Triticum polonicum, a tetraploid wheat subspecies first described by Linnaeus in 1753 and which was instrumental in the study of quantitative variation by early geneticists such as Biffen. We recently cloned P1, the gene responsible for its striking glume morphology. 1/n
Here is the @biorxivpreprint link and below I'll try to tell the story of why this is exciting. 2/n https://www.biorxiv.org/content/10.1101/2020.11.09.375154v2
Note that there is also some fascinating discoveries in the companion work by @jmdebernardi and colleagues in the Dubcovsky lab @UCSmallGrains; also in @biorxivpreprint https://www.biorxiv.org/content/10.1101/2020.12.01.405779v1 3/n
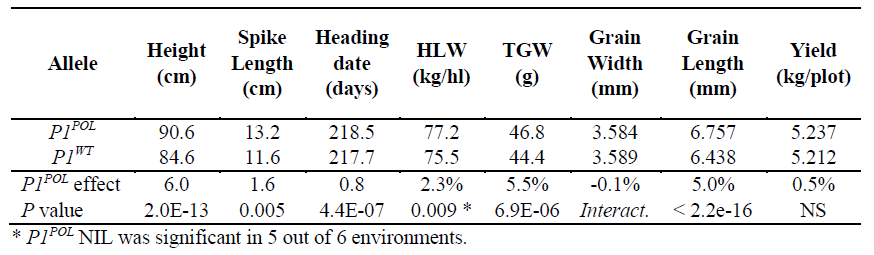
We developed near-isogenic lines of P1 into hexaploid wheat and found a significant increase in grain weight due to longer grains, and also test weight (i.e. the weight of grains which can be packed into a certain volume) (important for transport). however no effect on yield 4/n
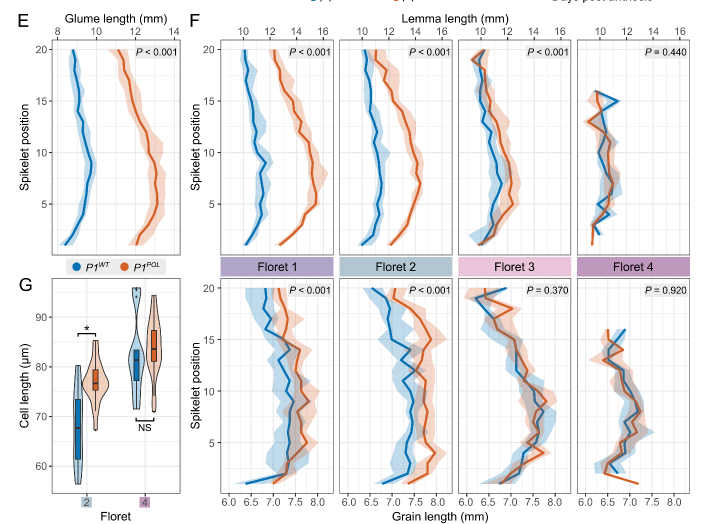
The NILs have longer glumes across the spike; however the effect on lemma length and grain length gradually decreases across the spikelet, with strongest phenotypic effects at the base of each spikelet. 5/n
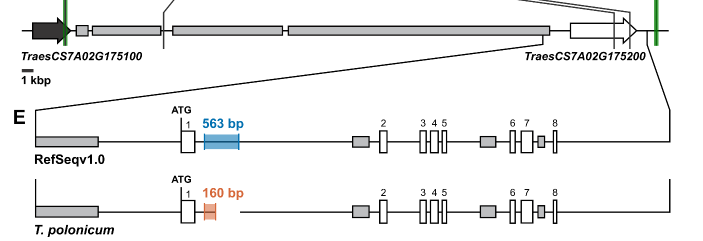
We mapped multiple P1 phenotypes (plant height, spike length, grain length and weight, glume length) to a single gene, VRT2. This gene is a MADS-box TF orthologous to Arabidopsis SVP genes . Interestingly, the T. polonicum allele carries an intron-1 re-arrangement. 6/n
We found that the T. polonicum 160-bp intron 1 was private to polonicum wheat. In hexaploid we found this allele in T. petropavlovskyi and Arrancada accessions which were previously proposed to be hybrids with polonicum. 7/n
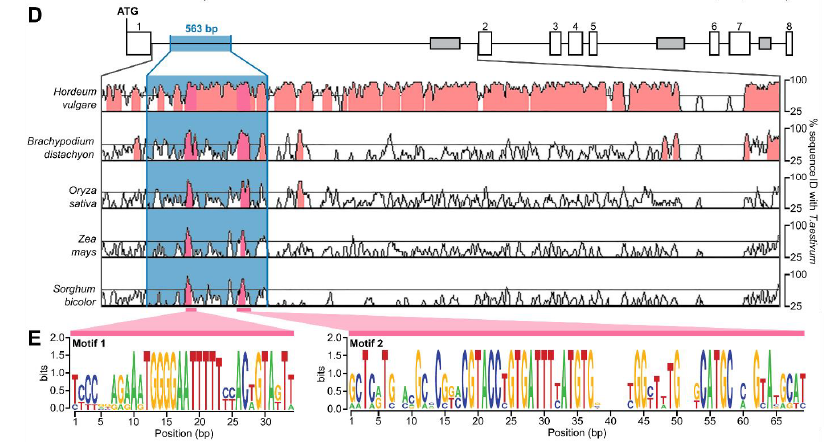
We looked at the wildtype 563-bp intron 1 sequence of VRT2 and found 2 sequences which are highly conserved across grasses, including maize and sorghum! 8/n
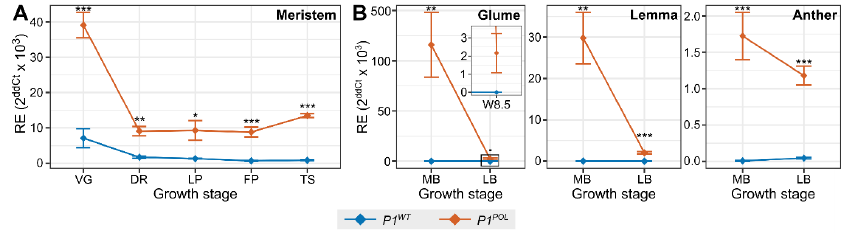
The 160-bp intron 1 sequence in T. polonicum is associated with higher and ectopic expression of VRT2. Normally VRT2 is downregulated in meristems and in T. polonicum we see that despite some downregulation, the VRT2 is expressed more highly and in new tissues glumes, etc. 9/n
We developed transgenic lines with the T. polonicum P1 allele and we show a dosage-dependent phenotypic response. High copy number lines have longer spikes, glumes and grains with respect to zero copy lines. 9/n
As always, all germplasm is freely available from the @JohnInnesCentre Germplasm Resources Unit (accession Code WM0013) https://www.seedstor.ac.uk/ . This allele is now being tested in different UK environments as part of @UKFutureWheat 10/n
Huge shout out to @NikolaiAdamski and @simmojsimmo who masterminded this project and have been steadily working on the genetics and all other aspects over the past 7 years. Incredible work by the team. Really proud. n=11

 Read on Twitter
Read on Twitter