Our study using @nanopore ONT sequencing to survey transposable element (TE) insertions and methylation landscapes in human tissues is now in print @MolecularCell. Detailed thread 
Open access link: https://www.sciencedirect.com/science/article/pii/S1097276520307310?dgcid=author
Work funded by @nhmrc @CSL @materfoundation
1/n

Open access link: https://www.sciencedirect.com/science/article/pii/S1097276520307310?dgcid=author
Work funded by @nhmrc @CSL @materfoundation
1/n
Included hippocampus, liver and heart for one person without pathology, and a tumour/non-tumour liver pair. Samples were ONT sequenced ~15x genome-wide and analysed with TLDR and other tools written here by @_adamewing. Observations below mostly for people interested in TEs.
2/n
2/n
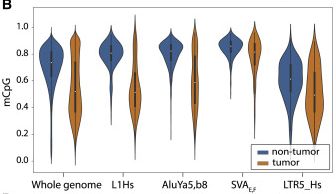
ONT sequencing is an incredible(!!) tool for looking at the methylation profiles of TEs genome-wide. For example, our ONT data from a liver tumour showed methylation on L1HS TE copies was far more reduced than for other TE types (e.g. SVA, Alu) or the genome overall.
3/n
3/n
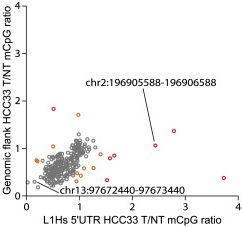
We found that, in cancer, L1HS methylation changes can appear strongly disjointed with that of the adjacent genome (higher OR lower), suggesting regulation specific to the L1HS, not just a passenger to genome-wide changes.
4/n
4/n
We previously found L1HS copies recurrently hypomethylated in cancer (e.g. https://genome.cshlp.org/content/28/5/639.short) and ONT analysis agrees with this. The tech is robust ... and many L1HS methylation changes in cancer are not random. The same L1HS copies keep popping up! TTC28 L1HS etc.
5/n
5/n
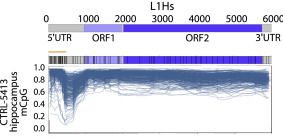
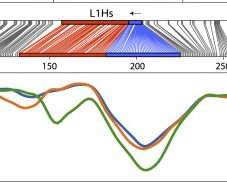
A major benefit of ONT sequencing is being able to fully resolve young TE methylation profiles. Here is L1HS family-wide methylation in hippocampus tissue, where each line represents an ONT read spanning >6kb. See the clear trough in the 5'UTR after the CpG island?
6/n
6/n
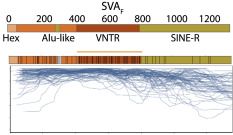
And here's the youngest SVA family in hippocampus tissue. SVA has a CpG island in its variable number of tandem repeats (VNTR) core and again we see methylation sloping away on either side of that CpG island.
7/n
7/n
ONT analysis clearly resolves methylation of individual TEs. Here, at left: an antisense chr13 L1HS mobile in the brain (hippocampus methylation blue). Right: bisulfite sequencing by @FranciscoJSL2 2019 paper for the first half of the L1HS 5'UTR in hippocampus. Concordant.
8/n
8/n
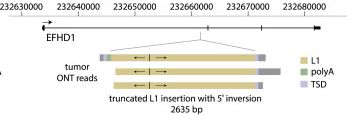
The ONT approach is great for finding and characterising new TE insertions. Below is a liver tumour somatic insertion previously found by us with Illumina sequencing (Shukla et al. 2013). It looks the same with ONT analysis but here we get the entire TE (TSDs etc) in one go
9/n
9/n
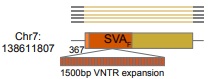
Long read ONT data can also reveal polymorphisms within young TE insertions. For example, SVA VNTR expansions seem to be common and are difficult to resolve with Illumina sequencing - not a problem for ONT sequencing. The below example has a 1.5kbp VNTR expansion!
10/n
10/n
All in all it was a fun project and one I enjoyed working on. Particular kudos to authors @_adamewing and @SethCheetham for instrumental contributions to concept and execution. Sincere thanks to the referees for their thoughtful input (Reviewer #1, whoever you are,  ).
).
11/11
 ).
). 11/11

 Read on Twitter
Read on Twitter